Identification of Pathogenic Fusarium spp. Causing Maize Ear Rot and Potential Mycotoxin Production in China
Abstract
:1. Introduction
2. Results
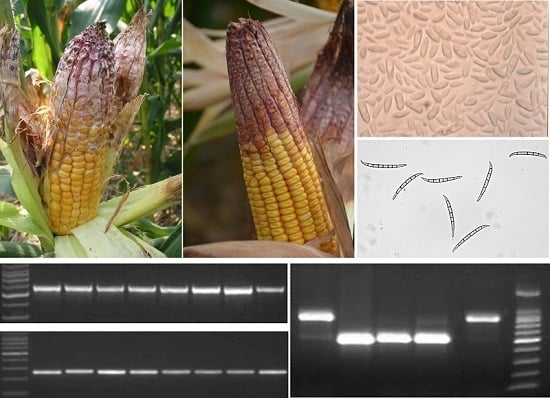
2.1. Identification of Fusarium spp.
2.2. Detection of Toxigenic Genes and Chemotypes
2.3. Detection of Toxigenic Capacity
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Isolation
4.2. Identification of Pathogenic Fungi
4.3. Molecular Identification of Toxigenic Genes
4.4. HPLC Detection of Mycotoxin Production
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 3-ADON | 3-acetyl-deoxynivalenol |
| 15-ADON | 15-deoxynivalenol |
| DON | deoxynivalenol |
| FB | fumonisin B |
| FGSC | Fusarium graminearum species complex |
| HPLC | high-performance liquid chromatography |
| LC-MS/MS | liquid chromatography-tandem mass spectrometry |
| ML | maximum likelihood |
| NIV | nivalenol |
| OPA | o-phthaldialdehyde |
| PCR | polymerase chain reactions |
| PDA | potato dextrose agar |
| SNA | Spezieller Nährstoffarmer agar |
| TEF | translation elongation factor |
| UHPLC-MS/MS | ultra-high performance liquid chromatography-mass spectrometry |
| ZEN | zearalenone |
References
- Görtz, A.; Oerke, E.C.; Steiner, U.; Waalwijk, C.; Vries, I.; Dehne, H.W. Biodiversity of Fusarium species causing ear rot of maize in Germany. Cereal Res. Commun. 2008, 36, 617–622. [Google Scholar] [CrossRef]
- Sampietro, D.A.; Diaz, C.G.; Gonzalez, V.; Vattuone, M.A.; Ploper, L.D.; Catalan, C.A.; Ward, T.J. Species diversity and toxigenic potential of Fusarium graminearum complex isolates from maize fields in northwest Argentina. Int. J. Food Microbiol. 2011, 145, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Covarelli, L.; Stifano, S.; Beccari, G.; Raggi, L.; Lattanzio, V.M.; Albertini, E. Characterization of Fusarium verticillioides strains isolated from maize in Italy: Fumonisin production, pathogenicity and genetic variability. Food Microbiol. 2012, 31, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.W.; Dyer, R.B.; McCormick, S.P.; Kendra, D.F.; Plattner, R.D. Functional demarcation of the Fusarium core trichothecene gene cluster. Fungal Genet. Biol. 2004, 41, 454–462. [Google Scholar] [CrossRef] [PubMed]
- Alexander, N.J.; Proctor, R.H.; McCormick, S.P. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium. Toxin Rev. 2009, 28, 198–215. [Google Scholar] [CrossRef]
- Yin, S.; Guo, X.; Li, J.; Fan, L.; Hu, H. Fumonisin B1 induces autophagic cell death via activation of ERN1-MAPK8/9/10 pathway in monkey kidney MARC-145 cells. Arch. Toxicol. 2016, 90, 985–996. [Google Scholar] [CrossRef] [PubMed]
- Gelderblom, W.C.; Jaskiewicz, K.; Marasas, W.F.; Thiel, P.G.; Horak, R.M.; Vleggaar, R.; Kriek, N.P. Fumonisins-novel mycotoxins with cancer-promoting activity produced by Fusarium moniliforme. Appl. Environ. Microbiol. 1998, 54, 1806–1811. [Google Scholar]
- Larsen, J.C.; Hunt, J.; Perrin, I.; Ruckenbauer, P. Workshop on trichothecenes with a focus on DON: Summary report. Toxicol. Lett. 2004, 153, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Aboul-Nasr, M.B.; Obied-Allah, M.R. Biological and chemical detection of fumonisins produced on agar medium by Fusarium verticillioides isolates collected from corn in Sohag, Egypt. Microbiology 2013, 159, 1720–1724. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Kersten, S.; Meyer, U.; Stinshoff, H.; Locher, L.; Rehage, J.; Wrenzycki, C.; Engelhardt, U.H.; Danicke, S. Diagnostic opportunities for evaluation of the exposure of dairy cows to the mycotoxins deoxynivalenol (DON) and zearalenone (ZEN): Reliability of blood plasma, bile and follicular fluid as indicators. J. Anim. Physiol. Anim. Nutr. 2015, 99, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Logrieco, A.; Mulè, G.; Moretti, A.; Bottalico, A. Toxigenic Fusarium species and mycotoxins associated with maize ear rot in Europe. Eur. J. Plant Pathol. 2002, 108, 597–609. [Google Scholar] [CrossRef]
- Zhang, X.F.; Zou, C.J.; Cui, L.N.; Li, X.; Yang, X.R.; Luo, H.H. Identification of pathogen causing maize ear rot and inoculation technique in Southwest China. Southwest China J. Agric. Sci. 2012, 25, 2078–2082. [Google Scholar]
- Qin, Z.H.; Ren, X.; Jiang, K.; Wu, X.F.; Yang, Z.H.; Wang, X.M. Identification of Fusarium species and F. graminearum species complex causing maize ear rot in China. Acta Phytophy. Sin. 2014, 41, 589–596. [Google Scholar]
- Fu, M.; Li, R.; Guo, C.; Pang, M.; Liu, Y.; Dong, J. Natural incidence of Fusarium species and fumonisins B1 and B2 associated with maize kernels from nine provinces in China in 2012. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2015, 32, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Rheeder, J.P.; Marasas, W.F.; Vismer, H.F. Production of fumonisin analogs by Fusarium species. Appl. Environ. Microbiol. 2002, 68, 2101–2105. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, T.T.; Zitomer, N.C.; Mitchell, T.R.; Zimeri, A.M.; Bacon, C.W.; Riley, R.T.; Glenn, A.E. Maize seedling blight induced by Fusarium verticillioides: Accumulation of Fumonisin B1 in leaves without colonization of the leaves. J. Agric. Food Chem. 2014, 62, 2118–2125. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.W.; Butchko, R.A.E.; Busman, M.; Proctor, R.H. The Fusarium verticillioides FUM gene cluster encodes a Zn(II)2Cys6 protein that affects FUM gene expression and fumonisin production. Eukaryot. Cell 2007, 6, 1210–1218. [Google Scholar] [CrossRef] [PubMed]
- Glenn, A.E.; Zitomer, N.C.; Zimeri, A.M.; Williams, L.D.; Riley, R.T.; Proctor, R.H. Transformation-mediated complementation of a FUM gene cluster deletion in Fusarium verticillioides restores both fumonisin production and pathogenicity on maize seedlings. Mol. Plant Microbe Interact. 2008, 21, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.H.; Plattner, R.D.; Desjardins, A.E.; Busman, M.; Butchko, R.A.E. Fumonisin production in the maize pathogen Fusarium verticillioides: Genetic basis of naturally occurring chemical variation. J. Agric. Food Chem. 2006, 54, 2424–2430. [Google Scholar] [CrossRef] [PubMed]
- Bottalico, A.; Perrone, G. Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. Eur. J. Plant Pathol. 2002, 108, 611–624. [Google Scholar] [CrossRef]
- Merhej, J.; Richard-Forget, F.; Barreau, C. Regulation of trichothecene biosynthesis in Fusarium: Recent advances and new insights. Appl. Microbiol. Biotechnol. 2011, 91, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.X.; Wang, X.M.; Song, F.J.; Sun, S.L.; Zhou, D.L.; Zhu, Z.D. Advances in research on maize resistance to ear rot. Sci. Agric. Sin. 2015, 48, 2152–2164. [Google Scholar]
- Pestka, J.J. Deoxynivalenol: Mechanisms of action, human exposure, and toxicological relevance. Arch. Toxicol. 2010, 84, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.D.; Greenhalgh, R.; Wang, Y.Z.; Lu, M. Trichothecene chemotypes of three Fusarium species. Mycologia 1991, 83, 121–130. [Google Scholar] [CrossRef]
- McCormick, S.P.; Stanley, A.M.; Stover, N.A.; Alexander, N.J. Trichothecenes: from simple to complex mycotoxins. Toxins 2011, 3, 802–814. [Google Scholar] [CrossRef] [PubMed]
- Burlakoti, R.R.; Estrada, R.; Rivera, V.V.; Boddeda, A.; Secor, G.A.; Adhikari, T.B. Real-time PCR quantification and mycotoxin production of Fusarium graminearum in wheat inoculated with isolates collected from potato, sugar beet, and wheat. Phytopathology 2007, 97, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Burlakoti, R.R.; Ali, S.; Secor, G.A.; Neate, S.M.; McMullen, M.P.; Adhikari, T.B. Comparative mycotoxin profiles of Gibberella zeae populations from barley, wheat, potatoes, and sugar beets. Appl. Environ. Microbiol. 2008, 74, 6513–6520. [Google Scholar] [CrossRef] [PubMed]
- Foroud, N.; MacMillan, T.; Ellis, B.; Eudes, F. The role of trichothecene-chemotypes in Fusarium head blight disease spread in wheat. Cereal Res. Commun. 2008, 36, 489–490. [Google Scholar]
- Ward, T.J.; Clear, R.M.; Rooney, A.P.; O’Donnell, K.; Gaba, D.; Patrick, S.; Starkey, D.E.; Gilbert, J.; Geiser, D.M.; Nowicki, T.W. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet. Biol. 2008, 445, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Jestoi, M. Emerging Fusarium-mycotoxins fusaproliferin, beauvericin, enniatins, and moniliformin—A review. Crit. Rev. Food Sci. Nutr. 2008, 48, 21–49. [Google Scholar] [CrossRef] [PubMed]
- Yli-Mattila, T.; Gagkaeva, T.; Ward, T.J.; Aoki, T.; Kistler, H.C.; O’Donnell, K. A novel Asian clade within the Fusarium graminearum species complex includes a newly discovered cereal head blight pathogen from the Russian Far East. Mycologia 2009, 101, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Yli-Mattila, T.; Gagkaeva, T. Molecular chemotyping of Fusarium graminearum, F. culmorum, and F. cerealis isolates from Finland and Russia. In Molecular Identification of Fungi; Gherbawy, Y., Voigt, K., Eds.; Springer Verlag: Berlin, Germany; Heidelberg, Germany; New York, NY, USA, 2010; pp. 159–177. [Google Scholar]
- Pasquali, M.F.; Brochot, C.; Cocco, E.; Hoffmann, L.; Bohn, T. Genetic Fusarium chemotyping as a useful tool for predicting nivalenol contamination in winter wheat. Int. J. Food Microbiol. 2010, 137, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Gang, G.; Miedaner, T.; Schuhmacher, U.; Schollenberger, M.; Geiger, H.H. Deoxynivalenol and nivalenol production by Fusarium culmorum isolates differing in aggressiveness toward winter rye. Phytopathology 1998, 88, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, M.; Beyer, M.; Logrieco, A.; Audenaert, K.; Balmas, V.; Basler, R.; Boutigny, A.L.; Chrpová, J.; Czembor, E.; Gagkaeva, T.; et al. A European database of Fusarium graminearum and F. culmorum trichothecene genotypes. Front. Microbiol. 2016, 7, 406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krska, R.; Baumgartner, S.; Josephs, R. The state of the art in the analysis of type-A and -B trichothecene mycotoxins in cereals. J. Anal. Chem. 2001, 371, 285–299. [Google Scholar] [CrossRef]
- Li, W.X.; Zheng, C.M.; Wu, L.; Li, X.; Li, J.M.; Song, J.K.; Yang, X.L.; Wang, B.J. Determining fumonisins in corn by high performance liquid chromatography with immunoaffinity column cleanup. Acta Agron. Sin. 2012, 38, 556–562. [Google Scholar] [CrossRef]
- Escobar, J.; Loran, S.; Gimenez, I.; Ferruz, E.; Herrera, M.; Herrera, A.; Arino, A. Occurrence and exposure assessment of Fusarium mycotoxins in maize germ, refined corn oil and margarine. Food Chem. Toxicol. 2013, 62, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Li, W.X.; Zhang, Y.; Sun, L.J.; Dong, X.L.; Hu, X.X.; Wang, B.J. Simultaneous determination of twelve mycotoxins in cereals by ultra-high performance liquid chromatography-tandem mass spectrometry. Acta Agron. Sin. 2014, 40, 691–701. [Google Scholar] [CrossRef]
- Guo, C.; Wei, H.Y.; Guo, M.K.; He, S.Q.; Jin, S.L.; Chen, H.M.; Wang, X.M.; Guo, J.G. Isolation, identification and biological characteristics of Fusarium verticillioides from maize ear rot samples in Gansu Province. Acta Phytopathol. Sin. 2014, 44, 17–25. [Google Scholar]
- Sanchèz-Rangel, D.; Sanjuan-Badillo, A.; Plasencia, J. Fumonisin production by Fusarium verticillioides strains isolated from maize in Mexico and development of a polymerase chain reaction to detect potential toxigenic strains in grains. J. Agric. Food Chem. 2005, 53, 8565–8571. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Ward, T.J.; Geiser, D.M.; Corby, K.H.; Aoki, T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 2004, 41, 600–623. [Google Scholar]
- Starkey, D.E.; Ward, T.J.; Aoki, T.; Gale, L.R.; Kistler, H.C.; Geiser, D.M.; Suga, H.; Toth, B.; Varga, J.; O’Donnell, K. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 2007, 44, 1191–1204. [Google Scholar] [CrossRef] [PubMed]
- Lamprecht, S.C.; Tewoldemedhin, Y.T.; Botha, W.J.; Calitz, F.J. Fusarium graminearum species complex associated with maize crowns and roots in the KwaZulu-Natal province of South Africa. Plant Dis. 2011, 95, 1153–1158. [Google Scholar] [CrossRef]
- Sugiura, Y.; Watanabe, Y.; Tanaka, T.; Yamamoto, S.; Ueno, Y. Occurrence of Gibberella zeae strains that produce both nivalenol and deoxynivalenol. Appl. Environ. Microbiol. 1990, 56, 3047–3051. [Google Scholar] [PubMed]
- Kim, H.S.; Lee, T.; Dawlatana, M.; Yun, S.H.; Lee, Y.W. Polymorphism of trichothecene biosynthesis genes in deoxynivalenol- and nivalenol-producing Fusarium graminearum isolates. Mycol. Res. 2003, 107, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; Blackwell: Iowa City, IA, USA, 2006. [Google Scholar]
- Liu, D.; Coloe, S.; Baird, R.; Pedersen, J. Rapid mini-preparation of fungal DNA for PCR. J. Clin. Microbiol. 2000, 38, 471. [Google Scholar] [PubMed]
- Bluhm, B.H.; Flaherty, J.E.; Cousin, M.A.; Woloshuk, C.P. Multiplex polymerase chain reaction assay for the differential detection of trichothecene- and fumonisin-producing species of Fusarium in cornmeal. J. Food Prot. 2002, 65, 1955–1961. [Google Scholar] [PubMed]
- Nicholson, P.; Simpson, D.R.; Weston, G.; Rezanoor, H.N.; Lees, A.K.; Parry, D.W.; Joyce, D. Detection and quantification of Fusarium culmorum and Fusarium graminearumin cereals using PCR assays. Physiol. Mol. Plant Pathol. 1998, 53, 17–37. [Google Scholar] [CrossRef]
- Mishra, P.K.; Fox, R.T.V.; Culham, A. Development of a PCR-based assay for rapid and reliable identification of pathogenic Fusariam. FEMS Microbiol. Lett. 2003, 218, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Mulè, G.; Susca, A.; Stea, G.; Moretti, A. A species-specific PCR assay based on the calmodulin partial gene for identification of Fusarium verticillioides, F. proliferatum and F. subglutinans. Eur. J. Plant Pathol. 2004, 110, 495–502. [Google Scholar] [CrossRef]
- Watanabe, M.; Yonezawa, T.; Lee, K.; Kumagai, S.; Sugita-Konishi, Y.; Goto, K.; Hara-Kudo, Y. Molecular phylogeny of the higher and lower taxonomy of the Fusarium genus and differences in the evolutionary histories of multiple genes. BMC Evol. Biol. 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Cyber-infrastructure for Fusarium. Available online: http://www.fusariumdb.org/index.php (accessed on 5 March 2014).
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Li, H.P.; Qu, B.; Zhang, J.B.; Huang, T.; Chen, F.F.; Liao, Y.C. Development of a generic PCR detection of 3-acetyldeoxy-nivalenol-, 15-acetyldeoxynivalenol- and nivalenol-chemotypes of Fusarium graminearum Clade. Int. J. Mol. Sci. 2008, 9, 2495–2504. [Google Scholar] [CrossRef] [PubMed]


| No. | Origin of Isolate | FB1 (µg/g) | FB2 (µg/g) | FB3 (µg/g) | FB (µg/g) |
|---|---|---|---|---|---|
| FVAH-1 | Sixian, Anhui | 3437.79 ± 39.21 | 253.12 ± 11.23 | 69.91 ± 2.66 | 3760.82 ± 51.21 |
| FVAH-2 | Wuhe, Anhui | 533.71 ± 15.32 | 133.42 ± 8.11 | 27.90 ± 3.58 | 695.04 ± 15.22 |
| FVAH-3 | Xiaoxian, Anhui | 695.72 ± 13.89 | 76.33 ± 6.23 | 30.18 ± 1.56 | 802.24 ± 13.56 |
| FVAH-4 | Sixian, Anhui | 4837.44 ± 56.66 | 248.03 ± 7.89 | 121.23 ± 8.44 | 5206.70 ± 56.67 |
| FVAH-5 | Suzhou, Anhui | 1427.11 ± 20.13 | 150.57 ± 5.24 | 31.67 ± 2.31 | 1609.35 ± 22.14 |
| FVAH-6 | Suzhou, Anhui | 1605.32 ± 23.25 | 170.88 ± 4.61 | 44.21 ± 2.85 | 1820.41 ± 20.41 |
| FVBJ-1 | Changping, Beijing | 123.00 ± 9.85 | 48.18 ± 2.12 | 8.00 ± 1.13 | 179.18 ± 13.01 |
| FVGS-1 | Zhenyuan, Gansu | 1750.53 ± 15.22 | 144.26 ± 3.43 | 25.77 ± 1.89 | 1920.56 ± 15.21 |
| FVGZ-1 | Bijie, Guizhou | 2432.10 ± 13.85 | 172.07 ± 12.88 | 27.01 ± 2.14 | 2631.18 ± 23.33 |
| FVGZ-2 | Xifeng, Guizhou | 14.06 ± 1.31 | 33.46 ± 2.56 | 24.51 ± 0.92 | 72.03 ± 5.85 |
| FVGZ-3 | Xifeng, Guizhou | 12.58 ± 1.02 | 30.12 ± 1.78 | 21.87 ± 088 | 64.57 ± 4.12 |
| FVGZ-4 | Qianxinan, Guizhou | 14.29 ± 1.15 | 31.27 ± 1.36 | 22.91 ± 3.11 | 68.47 ± 3.66 |
| FVHB-1 | Shenzhou, Hebei | 40.04 ± 2.89 | 14.10 ± 2.11 | 47.26 ± 0.98 | 101.39 ± 10.23 |
| FVHB-2 | Hengshui, Hebei | 226.85 ± 8.87 | 71.20 ± 5.36 | 10.25 ± 2.10 | 308.29 ± 11.25 |
| FVHB-3 | Cangzhou, Hebei | 102.81 ± 5.36 | 31.49 ± 2.88 | 5.63 ± 1.36 | 139.94 ± 12.03 |
| FVHB-4 | Cangzhou, Hebei | 123.15 ± 3.85 | 42.59 ± 2.56 | 7.67 ± 1.87 | 173.41 ± 9.87 |
| FVHB-5 | Yongnian, Hebei | 11.25 ± 1.39 | 27.95 ± 3.11 | 15.65 ± 2.58 | 54.85 ± 5.36 |
| FVHB-6 | Handan, Hebei | 5.81 ± 0.97 | 14.90 ± 1.01 | 9.10 ± 1.30 | 29.81 ± 3.99 |
| FVHB-7 | Qinhuangdao, Hebei | 405.76 ± 11.21 | 36.85 ± 1.23 | 6.71 ± 0.56 | 449.33 ± 8.23 |
| FVHB-8 | Luanxian, Hebei | 374.05 ± 8.89 | 32.78 ± 2.58 | 8.73 ± 0.55 | 415.55 ± 14.25 |
| FVHB-9 | Zhangjiakou, Hebei | 62.43 ± 3.25 | 23.44 ± 0.97 | 4.98 ± 0.87 | 90.85 ± 6.86 |
| FVHN-1 | Zhengzhou, Henan | 54.53 ± 2.55 | 0.00 | 3.47 ± 0.64 | 57.99 ± 7.11 |
| FVHN-2 | Zhengzhou, Henan | 350.58 ± 9.36 | 65.81 ± 3.12 | 8.00 ± 1.02 | 424.39 ± 13.58 |
| FVHN-3 | Zhongmo, Henan | 1072.13 ± 23.56 | 61.19 ± 2.58 | 42.43 ± 2.37 | 1175.75 ± 35.56 |
| FVHN-4 | Xunxian, Henan | 7.46 ± 0.91 | 8.03 ± 0.46 | 0.28 ± 0.04 | 15.77 ± 1.91 |
| FVHN-6 | Luoyang, Henan | 3195.88 ± 33.51 | 755.84 ± 6.35 | 56.30 ± 3.25 | 4008.02 ± 80.12 |
| FVHN-7 | Luoyang, Henan | 2886.75 ± 30.12 | 661.72 ± 5.01 | 50.12 ± 3.68 | 3598.59 ± 53.13 |
| FVHN-8 | Fangcheng, Henan | 280.79 ± 8.87 | 51.09 ± 1.25 | 5.83 ± 0.65 | 337.70 ± 23.14 |
| FVHN-9 | Luyi, Henan | 276.79 ± 6.84 | 98.93 ± 2.11 | 3.85 ± 0.27 | 379.56 ± 11.08 |
| FVHN-10 | Zhumadian, Henan | 13,456.63 ± 89.23 | 1864.90 ± 20.14 | 96.37 ± 3.87 | 15,417.91 ± 200.73 |
| FVHLJ-1 | Qiqihar, Heilongjiang | 2.98 ± 0.31 | 8.65 ± 0.63 | 2.69 ± 0.29 | 14.32 ± 0.86 |
| FVHLJ-2 | Qiqihar, Heilongjiang | 3.79 ± 0.41 | 10.25 ± 0.87 | 3.86 ± 0.27 | 17.90 ± 0.65 |
| FVHLJ-3 | Harbin, Heilongjiang | 992.17 ± 8.11 | 120.22 ± 10.54 | 18.24 ± 3.45 | 1130.63 ± 21.21 |
| FVHLJ-4 | Harbin, Heilongjiang | 3.62 ± 0.65 | 9.54 ± 0.68 | 6.16 ± 0.67 | 19.33 ± 1.22 |
| FVHLJ-5 | Suihua, Heilongjiang | 5140.81 ± 32.23 | 652.95 ± 9.35 | 50.84 ± 2.35 | 5844.59 ± 23.56 |
| FVHUB-1 | Enshi, Hubei | 21.54 ± 1.52 | 8.23 ± 0.21 | 16.87 ± 3.45 | 46.64 ± 2.24 |
| FVHUN-2 | Changsha, Hunan | 51.04 ± 1.88 | 12.05 ± 0.66 | 101.46 ± 12.03 | 164.56 ± 3.21 |
| FVJL-1 | Dehui, Jining | 18,416.44 ± 66.12 | 441.6818.11 | 356.31 ± 15.22 | 19,214.44 ± 159.32 |
| FVLN-1 | Youyan, Liaoning | 5.75 ± 0.89 | 11.38 ± 1.02 | 14.93 ± 1.28 | 32.05 ± 2.14 |
| FVLN-2 | Donggang, Liaoning | 0.00 | 0.00 | 0.00 | 0.00 |
| FVLN-3 | Fengcheng, Liaoning | 1243.47 ± 13.56 | 203.89 ± 11.25 | 66.77 ± 0.89 | 1514.13 ± 23.14 |
| FVLN-4 | Fengcheng, Liaoning | 1103.47 ± 11.33 | 198.35 ± 9.12 | 53.07 ± 0.45 | 1354.89 ± 19.88 |
| FVLN-5 | Fumeng, Liaoning | 281.38 ± 5.78 | 50.88 ± 3.11 | 150.28 ± 10.22 | 482.54 ± 11.25 |
| FVLN-6 | Fumeng, Liaoning | 18.47 ± 1.23 | 7.81 ± 0.78 | 14.35 ± 1.78 | 40.62 ± 3.32 |
| FVLN-7 | Fumeng, Liaoning | 3.84 ± 0.75 | 9.33 ± 0.56 | 4.39 ± 0.56 | 17.56 ± 2.11 |
| FVNMG-1 | Chifeng, Inner Mongolia | 586.63 ± 8.22 | 245.79 ± 11.23 | 55.16 ± 4.23 | 887.58 ± 12.01 |
| FVNMG-2 | Wuhai, Inner Mongolia | 45.01 ± 3.11 | 9.25 ± 0.88 | 44.30 ± 3.55 | 98.56 ± 12.37 |
| FVNX-1 | Tongxin, Ningxia | 11.74 ± 1.03 | 8.67 ± 0.97 | 9.34 ± 1.02 | 29.75 ± 3.21 |
| FVNX-2 | Yinchuan, Ningxia | 213.72 ± 8.47 | 0.00 | 8.69 ± 0.89 | 222.41 ± 22.17 |
| FVNX-3 | Yongning, Ningxia | 180.78 ± 3.66 | 39.89 ± 3.54 | 15.02 ± 1.58 | 235.68 ± 6.89 |
| FVSD-1 | Dezhou, Shandong | 0.00 | 0.00 | 0.00 | 0.00 |
| FVSD-2 | Dezhou, Shandong | 7.78 ± 1.12 | 6.60 ± 0.65 | 9.30 ± 1.07 | 23.68 ± 3.58 |
| FVSD-3 | Yuncheng, Shandong | 7.91 ± 0.76 | 5.55 ± 0.77 | 13.32 ± 0.56 | 26.77 ± 1.25 |
| FVSD-4 | Jiaxiang, Shandong | 3.55 ± 0.43 | 3.40 ± 0.61 | 1.04 ± 0.11 | 7.99 ± 0.81 |
| FVSD-5 | Jining, Shandong | 3.24 ± 0.77 | 5.39 ± 0.56 | 3.88 ± 0.57 | 12.51 ± 0.56 |
| FVSD-6 | Jining, Shandong | 5.35 ± 0.87 | 7.69 ± 0.86 | 5.93 ± 0.64 | 18.97 ± 0.47 |
| FVSD-7 | Sishui, Shandong | 88.43 ± 5.44 | 13.57 ± 1.57 | 61.29 ± 1.26 | 163.29 ± 23.57 |
| FVSD-8 | Weishan, Shandong | 4.14 ± 0.39 | 3.36 ± 0.67 | 5.26 ± 0.88 | 12.77 ± 1.87 |
| FVSD-9 | Yanzhou, Shandong | 2.52 ± 0.57 | 3.00 ± 0.25 | 3.24 ± 0.54 | 8.75 ± 0.65 |
| FVSD-10 | Liaocheng, Shandong | 204.66 ± 10.13 | 61.96 ± 3.31 | 11.95 ± 1.06 | 278.58 ± 12.37 |
| FVSD-11 | Pingdu, Shandong | 2.61 ± 0.13 | 402.81 ± 23.14 | 89.43 ± 5.33 | 494.85 ± 23.11 |
| FVSD-12 | Qingzhou, Shandong | 1196.33 ± 22.11 | 73.85 ± 11.01 | 321.18 ± 11.23 | 1591.37 ± 20.87 |
| FVSD-13 | Shouguang, Shandong, | 2405.54 ± 28.32 | 106.06 ± 5.23 | 1510.54 ± 46.31 | 4022.15 ± 42.56 |
| FVSD-14 | Laizhou, Shandong | 145.34 ± 8.46 | 25.82 ± 2.45 | 74.04 ± 8.12 | 245.20 ± 15.21 |
| FVS-1 | Changzhi, Shanxi | 4402.45 ± 25.32 | 555.28 ± 35.43 | 1123.88 ± 23.21 | 6081.61 ± 53.25 |
| FVSX-1 | Baoji, Shaanxi | 312.24 ± 12.78 | 18.66 ± 2.55 | 21.95 ± 3.01 | 352.85 ± 10.04 |
| FVSX-2 | Qishan, Shaanxi | 253.62 ± 13.33 | 0.00 | 13.25 ± 2.06 | 266.87 ± 16.23 |
| FVSX-3 | Danfeng, Shaanxi | 4384.86 ± 35.41 | 287.50 ± 5.37 | 61.46 ± 3.02 | 4733.82 ± 35.65 |
| FVSX-4 | Shangluo, Shaanxi | 3476.72 ± 30.12 | 351.97 ± 15.36 | 95.03 ± 3.11 | 3923.72 ± 40.24 |
| FVSX-5 | Shangluo, Shaanxi | 2486.72 ± 15.32 | 231.51 ± 9.31 | 65.13 ± 2.18 | 2783.36 ± 29.81 |
| FVSX-6 | Luonan, Shaanxi | 21.52 ± 1.13 | 12.06 ± 2.14 | 16.42 ± 1.56 | 50.00 ± 3.60 |
| FVSX-7 | Luonan, Shaanxi | 2383.63 ± 23.08 | 155.53 ± 6.87 | 310.49 ± 9.23 | 2849.66 ± 50.17 |
| FVSX-8 | Fuping, Shaanxi | 210.44 ± 5.63 | 56.57 ± 3.47 | 17.26 ± 1.25 | 284.27 ± 23.14 |
| FVSX-9 | Huxian, Shaanxi | 23.54 ± 2.13 | 0.00 | 0.00 | 23.54 ± 2.38 |
| FVSC-1 | Zhongjiang, Sichuan | 223.58 ± 15.26 | 0.00 | 0.00 | 223.58 ± 10.25 |
| FVSC-2 | Santai, Sichuan | 2.26 ± 0.23 | 3.63 ± 0.58 | 0.88 ± 0.05 | 6.77 ± 0.67 |
| FVSC-3 | Mianyang, Sichuan | 77.40 ± 2.36 | 0.00 | 0.00 | 77.40 ± 3.54 |
| FVSC-4 | Xichong, Sichuan | 17.73 ± 1.09 | 5.14 ± 0.82 | 28.86 ± 5.23 | 51.72 ± 3.14 |
| FVYN-1 | Mangshi, Yunnan | 72.25 ± 2.11 | 15.88 ± 1.82 | 111.17 ± 4.77 | 199.30 ± 8.12 |
| FVYN-2 | Dehong, Yunnan | 0.00 | 0.00 | 0.00 | 0.00 |
| FVYN-3 | Mile, Yunnan | 40.23 ± 5.21 | 0.00 | 0.00 | 40.23 ± 3.69 |
| FVYN-4 | Qujing, Yunnan | 114.54 ± 5.67 | 0.00 | 10.07 ± 0.78 | 124.61 ± 10.25 |
| No. | Species | Isolate of Origin | Chemotype | DON (µg/g) | 15-ADON (µg/g) | 3-ADON (µg/g) | ZEN (µg/g) |
|---|---|---|---|---|---|---|---|
| FG001 | F. g. | Mengcheng, Anhui | 15-ADON | 7392.35 ± 48.02 | 27,713.62 ± 181.02 | 8316.53 ± 86.32 | 0.00 |
| FG008 | F. g. | Shunyi, Beijing | 15-ADON | 304.06 ± 15.36 | 1920.07 ± 23.36 | 528.73 ± 23.21 | 46.31 ± 3.25 |
| FG012 | F. g. | Zhuanglang, Gansu | 15-ADON | 19,795.33 ± 99.23 | 4350.49 ± 35.14 | 1315.46 ± 35.25 | 6.95 ± 0.81 |
| FG023 | F. g. | Changli, Hebei | 15-ADON | 4319.20 ± 35.62 | 24,411.32 ± 150.12 | 8167.15 ± 78.23 | 0.00 |
| FG029 | F. g. | Zhangjiakou, Hebei | 15-ADON | 12,464.99 ± 88.25 | 33,162.16 ± 223.57 | 11,471.99 ± 113.14 | 198.96 ± 13.37 |
| FG030 | F. g. | Tangshan, Hebei | 15-ADON | 3898.09 ± 46.35 | 18,546.56 ± 88.17 | 5674.04 ± 53.62 | 0.00 |
| FG039 | F. g. | Xiangcheng, Henan | 15-ADON | 0.00 | 0.00 | 0.00 | 0.00 |
| FG042 | F. g. | Sanmenxia, Henan | 15-ADON | 3189.00 ± 23.56 | 18,175.55 ± 100.25 | 5297.00 ± 85.36 | 0.00 |
| FG043 | F. g. | Zhoukou, Henan | 15-ADON | 394.64 ± 12.23 | 5956.24 ± 65.36 | 1863.57 ± 36.52 | 0.00 |
| FG044 | F. g. | Zhumadian, Henan | 15-ADON | 7955.22 ± 65.57 | 34,732.64 ± 137.58 | 9981.63 ± 89.65 | 0.00 |
| FG050 | F. g. | Qiqihar, Heilongjiang | 15-ADON | 4267.29 ± 23.39 | 6453.04 ± 82.21 | 1797.64 ± 13.56 | 0.00 |
| FG060 | F. g. | Donggang, Liaoning | 15-ADON | 406.97 ± 12.35 | 6967.08 ± 56.34 | 1718.66 ± 56.23 | 0.00 |
| FG063 | F. g. | Shenyang, Liaoning | 15-ADON | 1757.88 ± 21.45 | 4615.86 ± 58.75 | 1328.25 ± 33.21 | 2.73 ± 0.51 |
| FG072 | F. g. | Pingdu, Shandong | 15-ADON | 3172.77 ± 39.23 | 12,843.81 ± 98.21 | 3430.94 ± 25.24 | 6.18 ± 0.39 |
| FG075 | F. g. | Jiaxiang, Shandong | 15-ADON | 685.04 ± 15.63 | 10,420.73 ± 89.45 | 2869.32 ± 33.21 | 55.57 ± 3.21 |
| FG078 | F. g. | Dezhou, Shandong | 15-ADON | 8566.56 ± 46.36 | 54,382.05 ± 256.32 | 17,855.15 ± 78.56 | 6.83 ± 1.02 |
| FG080 | F. g. | Liaocheng, Shandong | 15-ADON | 224.59 ± 22.01 | 857.22 ± 10.58 | 242.27 ± 11.24 | 0.00 |
| FG081 | F. g. | Liaocheng, Shandong | 15-ADON | 1308.70 ± 63.70 | 9308.46 ± 56.32 | 2934.81 ± 55.21 | 22.72 ± 0.97 |
| FG082 | F. g. | Tai`an, Shandong | 15-ADON | 0.00 | 72.99 ± 6.38 | 23.20 ± 2.12 | 0.00 |
| FG083 | F. g. | Yanzhou, Shandong | 15-ADON | 213.08 ± 13.54 | 1704.10 ± 20.12 | 451.66 ± 10.53 | 0.00 |
| FG084 | F. g. | Qingdao, Shandong | 15-ADON | 0.00 | 25.52 ± 2.58 | 6.04 ± 0.65 | 0.00 |
| FG086 | F. g. | Laizhou, Shandong | 15-ADON | 1419.21 ± 21.08 | 7742.05 ± 33.56 | 1956.23 ± 21.32 | 0.00 |
| FG087 | F. g. | Weishan, Shandong | 15-ADON | 2417.01 ± 22.01 | 11,843.94 ± 87.58 | 2677.59 ± 35.26 | 0.00 |
| FG096 | F. g. | Xianyang, Shaanxi | 15-ADON | 1201.27 ± 36.25 | 8910.50 ± 98.24 | 2572.49 ± 63.27 | 0.00 |
| FG098 | F. g. | Fengxiang, Shaanxi | 15-ADON | 13.35 ± 1.04 | 86.84 ± 5.63 | 9.51 ± 0.59 | 0.00 |
| FG099 | F. g. | Yulin, Shaanxi | 15-ADON | 0.00 | 47.01 ± 6.23 | 10.14 ± 0.26 | 0.00 |
| FG101 | F. g. | Shangnan, Shaanxi | 15-ADON | 4764.23 ± 55.36 | 20,461.49 ± 100.58 | 5694.38 ± 66.39 | 0.00 |
| FG103 | F. g. | Zhashui, Shaanxi | 15-ADON | 2710.03 ± 12.37 | 8006.75 ± 89.57 | 2286.71 ± 55.31 | 0.00 |
| FG135 | F. g. | Sanmenxia, Henan | 15-ADON | 866.30 ± 26.31 | 4106.67 ± 76.35 | 1164.43 ± 53.37 | 1.77 ± 0.55 |
| FG141 | F. g. | Heishan, Liaoning | 15-ADON | 1668.64 ± 33.10 | 9666.53 ± 100.57 | 2425.36 ± 51.98 | 0 |
| FG144 | F. g. | Xianfeng, Hubei | 15-ADON | 5915.73 ± 29.25 | 26,784.49 ± 85.69 | 6497.77 ± 88.56 | 0.00 |
| FG144 | F. g. | Heihe, Helongjiang | 15-ADON | 0 | 174.24 ± 12.35 | 27.75 ± 3.04 | 3.63 ± 0.34 |
| FG147 | F. g. | Wangkui, Helongjiang | 15-ADON | 170.41 ± 8.63 | 311.93 ± 23.21 | 69.48 ± 3.56 | 88.27 ± 2.78 |
| FG150 | F. g. | Jiamusi, Helongjiang | 15-ADON | 12,422.55 ± 119.65 | 16,823.61 ± 88.56 | 4803.78 ± 40.12 | 0 |
| FG153 | F. g. | Fujin, Helongjiang | 15-ADON | 4742.29 ± 63.21 | 23,219.59 ± 150.69 | 6518.67 ± 45.23 | 0 |
| FG155 | F. g. | Fujin, Helongjiang | 15-ADON | 1825.81 ± 25.78 | 8084.01 ± 80.23 | 2064.13 ± 35.55 | 0 |
| FG156 | F. g. | Fengcheng, Liaoning | 15-ADON | 507.41 ± 13.69 | 2925.37 ± 23.01 | 575.05 ± 10.27 | 0 |
| FG158 | F. g. | Panshi, Jilin | 15-ADON | 16,723.72 ± 87.56 | 81,539.49 ± 300.57 | 19,590.61 ± 211.25 | 0 |
| FG159 | F. g. | Panshi, Jilin | 15-ADON | 698.51 ± 4.68 | 2845.5 ± 23.45 | 887.61 ± 6.38 | 0 |
| FG162 | F. g. | Nenjiang, Helongjiang | 15-ADON | 929.66 ± 15.23 | 5251.46 ± 23.22 | 1298.79 ± 37.24 | 14.37 ± 1.39 |
| FG165 | F. g. | Qinggang, Helongjiang | 15-ADON | 2998.93 ± 23.24 | 23,152.65 ± 123.5 | 5391.17 ± 52.13 | 430.24 ± 25.87 |
| FG015 | F. m. | Puding, Guizhou | NIV | 0.00 | 5.43 ± 0.56 | 17.24 ± 1.66 | 0.00 |
| FG017 | F. m. | Bijie, Guizhou | NIV | 0.00 | 0.00 | 0.00 | 0.00 |
| FG019 | F. m. | Guiyang, Guizhou | NIV | 0.00 | 20.46 ± 1.23 | 470.83 ± 12.25 | 0.00 |
| FG020 | F. m. | Zunyi, Guizhou | NIV | 0.00 | 28.82 ± 1.21 | 151.29 ± 13.24 | 0.00 |
| FG100 | F. m. | Luonan, Shaanxi | NIV | 0.00 | 67.44 ± 2.25 | 709.58 ± 33.22 | 0.00 |
| FG102 | F. m. | Danfeng, Shaanxi | 15-ADON, NIV | 0.00 | 12.29 ± 2.58 | 13.85 ± 1.65 | 0.00 |
| FG104 | F. m. | Zhenan, Shaanxi | 15-ADON | 69.60 ± 6.87 | 222.33 ± 11.012 | 63.93 ± 3.77 | 0.00 |
| FG105 | F. m. | Shangluo, Shaanxi | 15-ADON, NIV | 0.00 | 47.25 ± 3.12 | 53.13 ± 5.20 | 0.00 |
| FG106 | F. m. | Huayin, Shaanxi | NIV | 0.00 | 55.75 ± 1.25 | 767.60 ± 45.31 | 2.11 ± 0.49 |
| FG107 | F. m. | Luonan, Shaanxi | NIV | 0.00 | 40.05 ± 3.24 | 0.00 | 0.00 |
| FG128 | F. m. | Qujing, Yunnan | NIV | 0.00 | 8.58 ± 0.54 | 0.00 | 0.00 |
| FG129 | F. m. | Qujing, Yunnan | NIV | 0.00 | 11.46 ± 0.89 | 284.86 ± 23.56 | 8.90 ± 0.87 |
| FG130 | F. m. | Mile, Yunnan | NIV | 0.00 | 14.24 ± 1.55 | 78.62 ± 9.39 | 0.00 |
| FG133 | F. m. | Luxi, Yunnan | NIV | 0.00 | 104.90 ± 9.25 | 3519.87 ± 33.21 | 0.00 |
| FG140 | F. m. | Yuxi, Yunnan | NIV | 0.00 | 0.00 | 0.00 | 0.00 |
| FG142 | F. m. | Badong, Hubei | NIV | 0.00 | 0.00 | 0.00 | 0.00 |
| FG143 | F. m. | Jianshi, Hubei | NIV | 0.00 | 0.00 | 0.00 | 0.00 |
| FG145 | F. m. | Xianfeng, Hubei | NIV | 0.00 | 0.00 | 0.00 | 0.00 |
| FG146 | F. m. | Pixian, Sichuan | NIV | 0.00 | 0.00 | 0.00 | 0.00 |
| FG151 | F. m. | Mianyang, Sichuan | 15-ADON | 0.00 | 0.00 | 0.00 | 0.00 |
| FG152 | F. m. | Jianyang, Sichuan | 15-ADON | 0.00 | 0.00 | 0.00 | 0.00 |
| FG010 | F. b. | Haidian, Beijng | 15-ADON | 371.46 ± 12.56 | 2876.36 ± 36.56 | 843.70 ± 23.10 | 10.64 ± 1.11 |
| FG028 | F. b. | Zhangjiakou, Hebei | 15-ADON | 3636.37 ± 46.38 | 13,845.04 ± 95.63 | 4475.88 ± 45.32 | 0.00 |
| FG057 | F. b. | Huinan, Jilin | 15-ADON | 6794.02 ± 45.57 | 6844.28 ± 98.76 | 2132.45 ± 33.12 | 17.39 ± 0.97 |
| FG069 | F. b. | Chifeng, Inner Mongolia | 15-ADON | 9439.56 ± 80.67 | 9901.51 ± 85.69 | 2795.33 ± 46.29 | 0.00 |
| FG092 | F. b. | Changzhi, Shanxi | 15-ADON | 5629.31 ± 36.87 | 5687.94 ± 36.54 | 1779.75 ± 30.01 | 53.51 ± 3.24 |
| FG094 | F. b. | Changzhi, Shanxi | 15-ADON | 9939.69 ± 55.36 | 11,725.34 ± 111.30 | 3457.11 ± 55.32 | 0.00 |
| FG142 | F. b. | Suihua, Helongjiang | 15-ADON | 1491.91 ± 23.25 | 4549.23 ± 56.68 | 1334.72 ± 23.65 | 14.25 ± 2.16 |
| FG145 | F. b. | Tongliao, Inner Mongolia | 15-ADON | 3151.85 ± 53.23 | 7899.02 ± 36.89 | 2168.33 ± 33.45 | 0 |
| FG146 | F. b. | Tongliao, Inner Mongolia | 15-ADON | 0 | 38.83 ± 3.87 | 0 | 0 |
| FG147 | F. b. | Chengdu, Sichuan | 15-ADON | 0.00 | 0.00 | 0.00 | 0.00 |
| FG148 | F. b. | Liangshan, Sichuan | 15-ADON | 1156.47 ± 23.15 | 22123.45 ± 78.65 | 5838.04 ± 39.87 | 0.00 |
| FG149 | F. b. | Qiqihar, Helongjiang | 15-ADON | 1548.25 ± 35.24 | 6991.64 ± 36.56 | 2114.88 ± 24.21 | 60.05 ± 3.25 |
| FG160 | F. b. | Lanxi, Helongjiang | 15-ADON | 4236.92 ± 23.47 | 18455.36 ± 100.24 | 4741.94 ± 55.12 | 36.92 ± 3.67 |
| FG161 | F. b. | Qinggang, Helongjiang | 15-ADON | 518.48 ± 12.25 | 5451.17 ± 85.63 | 1408.57 ± 23.23 | 74.67 ± 5.26 |
| FG166 | F. b. | Zhenlai, Jilin | 15-ADON | 1496.17 ± 22.01 | 5324.07 ± 83.21 | 1386.40 ± 23.87 | 22.68 ± 2.98 |
| FG167 | F. b. | Huachuan, Helongjiang | 15-ADON | 14.20 ± 1.25 | 236.46 ± 23.56 | 153.75 ± 9.68 | 29.61 ± 2.65 |
| Province | Number of Counties | Province | Number of Counties |
|---|---|---|---|
| Anhui | 5 | Inner mongoria | 3 |
| Beijing | 2 | Jilin | 4 |
| Gansu | 2 | Liaoning | 5 |
| Guizhou | 6 | Ningxia | 3 |
| Hebei | 12 | Shaanxi | 14 |
| Heilongjiang | 8 | Shandong | 13 |
| Henan | 11 | Shanxi | 4 |
| Hubei | 4 | Sichuan | 7 |
| Hunan | 1 | Yunnan | 6 |
| Fungus | Primer | Sequence (5′–3′) | Target Fragment (bp) | Tm (°C) | Reference |
|---|---|---|---|---|---|
| Fusarium spp. | ItsF | AACTCCCAAACCCCTGTGAACATA | 431 | 58 | [49] |
| ItsR | TTTAACGGCGTGGCCGC | ||||
| F. graminearum | Fg16NF | ACAGATGACAAGATTCAGGCACA | 280 | 57 | [50] |
| Fg16NR | TTCTTTGACATCTGTTCAACCCA | ||||
| F. culmorum | Fc01F | ATGGTGAACTCGTCCTGGC | 570 | 59 | [50] |
| Fc01R | CCCTTCTTACGCCAATCTCG | ||||
| F. oxysporum | FoF1 | ACATACCACTTGTTGCCTCG | 340 | 58 | [51] |
| FoR1 | CGCCAATCAATTTGAGGAACG | ||||
| F. verticillioides | VER1 | CTTCCTGCGATGTTTCTCC | 578 | 56 | [52] |
| VER2 | AATTGGCCATTGGTATTATATATCTA | ||||
| F. proliferatum | PRO1 | CTTTCCGCCAAGTTTCTTC | 585 | 56 | [52] |
| PRO2 | TGTCAGTAACTCGACGTTGTTG | ||||
| F. subglutinans | SUB1 | CTGTCGCTAACCTCTTTATCCA | 631 | 56 | [52] |
| SUB2 | CAGTATGGACGTTGGTATTATATCTAA |
| Taxon | Collection Strain No. | Accession No. | Host | Geographic Origin |
|---|---|---|---|---|
| F. graminearum | VI 01028 | AJ543588.1 | Oat | Norway |
| F. meridionale | 28723 | AF212436.1 | Corn | Nepal |
| F. boothii | 26916 | AF212444.1 | Corn | South Africa |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, C.; Qin, Z.; Yang, Z.; Li, W.; Sun, S.; Zhu, Z.; Wang, X. Identification of Pathogenic Fusarium spp. Causing Maize Ear Rot and Potential Mycotoxin Production in China. Toxins 2016, 8, 186. https://doi.org/10.3390/toxins8060186
Duan C, Qin Z, Yang Z, Li W, Sun S, Zhu Z, Wang X. Identification of Pathogenic Fusarium spp. Causing Maize Ear Rot and Potential Mycotoxin Production in China. Toxins. 2016; 8(6):186. https://doi.org/10.3390/toxins8060186
Chicago/Turabian StyleDuan, Canxing, Zihui Qin, Zhihuan Yang, Weixi Li, Suli Sun, Zhendong Zhu, and Xiaoming Wang. 2016. "Identification of Pathogenic Fusarium spp. Causing Maize Ear Rot and Potential Mycotoxin Production in China" Toxins 8, no. 6: 186. https://doi.org/10.3390/toxins8060186
APA StyleDuan, C., Qin, Z., Yang, Z., Li, W., Sun, S., Zhu, Z., & Wang, X. (2016). Identification of Pathogenic Fusarium spp. Causing Maize Ear Rot and Potential Mycotoxin Production in China. Toxins, 8(6), 186. https://doi.org/10.3390/toxins8060186