Chemical Identity of Interaction of Protein with Reactive Metabolite of Diosbulbin B In Vitro and In Vivo
Abstract
:1. Introduction
2. Results
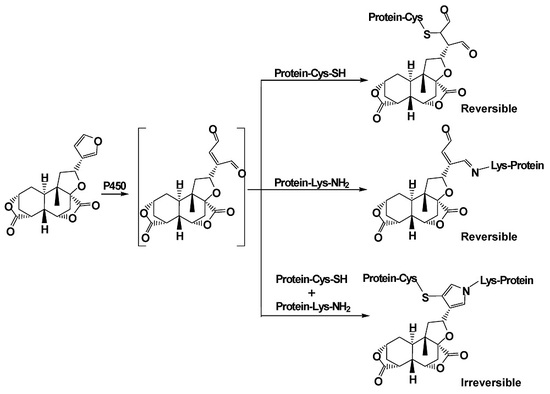
2.1. Cys/Lys Adduction by Reactive Intermediate of DIOB
2.2. Cys- and Lys-Based Protein Adduction by Reactive Metabolite of DIOB
2.3. Cys-Lys Based Protein Crosslink Derived from Bioactivation of DIOB
2.4. Reversibility of Protein Adduction by Reactive Metabolite of DIOB
2.5. Effect of P450 3A Activity on Microsomal Protein Adduction Induced by DIOB
2.6. Time- and Dose-Dependent Hepatic Protein Adduction Induced by DIOB
2.7. Effects of KTC and BSO on DIOB-Induced Hepatic Protein Adduction In Vivo
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Animals and Preparation of Liver Microsomes
4.3. Reactive Metabolite Trapping
4.4. Protein Adduction and Digestion
4.5. Dialysis Study
4.6. Chemical Synthesis
4.7. Animal Experiments
4.8. LC-MS/MS Method
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BSO | buthionine sulfoximine |
| BBA | 4-bromobenzylamine |
| BBM | 4-bromobenzylmercaptan |
| DIOB | diosbulbin B |
| DB | Dioscorea bulbifera L. |
| DEX | dexamethasone |
| CE | collision energy |
| HPLC/Q-TOF MS | high performance liquid chromatography/quadrupole-time-of-flight mass spectrometry |
| CXP | cell exit potential |
| DP | declustering potential |
| EP | entrance potential |
| EPI | enhanced product ion |
| IDA | information-dependent acquisition |
| KTC | ketoconazole |
| MRM | multiple-reaction monitoring |
| PI | precursor ion |
References
- Gao, H.; Kuroyanagi, M.; Wu, L.; Kawahara, N.; Yasuno, T.; Nakamura, Y. Antitumorpromoting constituents from Dioscorea bulbifera L. in JB6 mouse epidermal cells. Biol. Pharm. Bull. 2002, 25, 1241–1243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yuan, J. The progress of diosbulbin B research on pharmacology and Toxicology. Herald Med. 2009, 28, 490–492. [Google Scholar]
- Tang, Y. The research of Dioscoreae bulbifera L. in clinical application. Chin. J. Chin. Mater. Med. 1995, 20, 435–438. [Google Scholar]
- Rasikari, H.L.; Leach, D.N.; Waterman, P.G.; Spooner-Hart, R.N.; Basta, A.H.; Banbury, L.K.; Winter, K.M.; Forster, P.I. Cytotoxic clerodanediterpenes from Glossocaryacalcicola. Phytochemistry 2005, 66, 2844–2850. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Iliya, I.A.; Deng, J.; Zhao, S. Flavonoids and anthraquinone from Dioscorea bulbifera L. Chin. J. Chin. Mater. Med. 2000, 25, 159–160. [Google Scholar]
- Gao, H.; Shui, A.; Chen, Y.; Zhang, X.; Wu, L. The chemical compositions of Dioscorea bulbifera L. J. Shenyang Pharm. Univ. 2003, 20, 178–180. [Google Scholar]
- Grynberg, N.F.; Echevarria, A.; Lima, J.E.; Pamplona, S.S.; Pinto, A.C.; Maciel, M.A. Antitumour activity of two 19-nor-clerodane diterpenes, trans-dehydrocrotonin and trans-crotonin, from Croton cajucara. Planta Med. 1999, 65, 687–689. [Google Scholar] [CrossRef] [PubMed]
- Cifuente, D.A.; Borkowski, E.J.; Sosa, M.E.; Gianello, J.C.; Giordano, O.S.; Tonn, C.E. Clerodanediterpenes from Baccharissagittalis: Insect antifeedant activity. Phytochemistry 2002, 61, 899–905. [Google Scholar] [CrossRef]
- Demetzos, C.; Dimas, K.; Hatziantoniou, S.; Anastasaki, T.; Angelopoulou, D. Cytotoxic and anti-inflammatory activity of labdane and cis-clerodane type diterpenes. Planta Med. 2001, 67, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Teponno, R.B.; Tapondjou, A.L.; Gatsing, D.; Djoukeng, J.D.; Abou-Mansour, E.; Tabacchi, R.; Tane, P.; Stoekli-Evans, H.; Lontsi, D. Bafoudiosbulbins A, and B, two anti-salmonellal clerodane diterpenoids from Dioscorea bulbifera L. var sativa. Phytochemistry 2006, 67, 1957–1963. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.; Chen, A. 16 cases report of toxic hepatitis caused by Dioscorea bulbifera. Chin. J. Integr. Tradit. West. Liver Dis. 1994, 4, 55–56. [Google Scholar]
- Yang, H.; Li, J.; Cui, X.; Yang, C.; Li, L.; Liu, J. Clinical use and adverse drug reaction of compound prescription of Dioscorea bulbifera L. in clinical trial. Clin. Misdiagn. Misther. 2006, 19, 85–87. [Google Scholar]
- Liu, J. Two cases of toxic hepatitis caused by Dioscorea bulbifera L. Advers. Drug React. 2002, 2, 129–130. [Google Scholar]
- Wang, J.; Liang, Q.; Ji, L.; Liu, H.; Wang, C.; Wang, Z. Gender-related difference in liver injury induced by Dioscorea. bulbifera L. rhizome in mice. Hum. Exp. Toxicol. 2010, 30, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Niu, C.; Wang, J.; Ji, L.; Wang, Z. Diosbulbin B-induced liver injury in mice and its mechanism. Hum. Exp. Toxicol. 2013, 33, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lin, D.; Gao, H.; Xu, Y.; Meng, D.; Smith, C.V.; Peng, Y.; Zheng, J. Metabolic activation of furan moiety makes diosbulbin B hepatotoxic. Arch. Toxicol. 2016, 90, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Byrns, M.C.; Predecki, D.P.; Peterson, L.A. Characterization of nucleoside adducts of cis-2-butene-1,4-dial, a reactive metabolite of furan. Chem. Res. Toxicol. 2002, 15, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hecht, S.S.; Peterson, L.A. Characterization of amino acid and glutathione adducts of cis-2-butene-1,4-dial, a reactive metabolite of furan. Chem. Res. Toxicol. 1997, 10, 866–874. [Google Scholar] [CrossRef] [PubMed]
- Gates, L.A.; Lu, D.; Peterson, L.A. Trapping of cis-2-butene-1,4-dial to measure furan metabolism in human liver microsomes by cytochrome P450 enzymes. Drug Metab. Dispos. 2012, 40, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Peterson, L.A. Identification of furan metabolites derived from cysteine cis-2-butene-1,4-dial-lysine crosslinks. Chem. Res. Toxicol. 2010, 23, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Li, C.; Peng, Y.; Gao, H.; Zheng, J. Cytochrome P450-mediated metabolic activation of diosbulbin B. Drug. Metab. Dispos. 2014, 42, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lin, D.; Gao, H.; Hua, H.; Peng, Y.; Zheng, J. N-Acetyl lysine/glutathione-derived pyrroles as potential Ex Vivo biomarkers of bioactivated furan-containing compounds. Chem. Res. Toxicol. 2015, 28, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, W.; Chen, K.; Wang, Z.; Wang, C. Metabolism of diosbulbin B in vitro and in vivo in rats: Formation of reactive metabolites and human enzymes involved. Drug Metab. Dispos. 2015, 42, 1737–1750. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.B.; Sullivan, M.M.; Villalta, P.W.; Peterson, L.A. Covalent modification of cytochrome c by reactive metabolites of furan. Chem. Res. Toxicol. 2013, 27, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Moro, S.; Chipman, J.K.; Antczak, P.; Turan, N.; Dekant, W.; Falciani, F.; Mally, A. Identification and pathway mapping of furan target proteins reveal mitochondrial energy production and redox regulation as critical targets of furan toxicity. Toxicol. Sci. 2012, 126, 336–352. [Google Scholar] [CrossRef] [PubMed]
- Khojasteh, S.C.; Hartley, D.P.; Ford, K.A.; Uppal, H.; Oishi, S.; Nelson, S.D. Characterization of rat liver proteins adducted by reactive metabolites of menthofuran. Chem. Res. Toxicol. 2012, 25, 2301–2309. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, W.; Chen, J.; Peng, Y.; Zheng, J. Detection of cysteine- and lysine-based protein adductions by reactive metabolites of 2,5-dimethylfuran. Anal. Chim. Acta 2015, 896, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zheng, L.; Peng, Y.; Song, J.; Zheng, J. Selective and sensitive platform for function-based screening of potentially harmful furans. Anal. Chem. 2014, 86, 10755–10762. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Komori, T.; Setoguchi, S. Furanoidnorditerpenes from Dioscoreacae plants. 1. Diosbulins A, B, and C from Dioscorea. bulbifera form a spontanea. Chem. Pharm. Bull. 1968, 16, 2430–2435. [Google Scholar] [CrossRef]
- Lin, G.; Tang, J.; Liu, X.; Jiang, Y.; Zheng, J. Deacetylclivorine: A gender-selective metabolite of clivorine formed in female Sprague-Dawley rat liver microsomes. Drug Metab. Dispos. 2007, 35, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Li, L.; Zhong, D.; Liu, J.; Chen, X.; Zheng, J. Pulmonary toxicity and metabolic activation of tetrandrine in CD-1 mice. Chem. Res. Toxicol. 2011, 24, 2142–2152. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Shen, S.; Chen, X.; Zhong, D.; Zheng, J. CYP3A-mediated apoptosis of dauricine in cultured human bronchial epithelial cells and in lungs of CD-1 mice. Toxicol. Appl. Pharmacol. 2012, 261, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Zhang, H.; Jushchyshyn, M.; Hollenberg, P.F. Covalent modification of Thr302 in cytochrome P450 2B1 by the mechanism-based inactivator 4-tertbutylphenylacetylene. J. Pharmacol. Exp. Ther. 2010, 333, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Aloysius, H.; Tong, V.W.; Yabut, J.; Bradley, S.A.; Shang, J.; Zou, Y.; Tschirret-Guth, R.A. Metabolic activation and major protein target of a 1-benzyl-3-carboxyazetidine sphingosine-1-phosphate-1 receptor agonist. Chem. Res. Toxicol. 2012, 25, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Taber, D.F.; DeMatteo, P.W.; Hassan, R.A. Simplified Preparation of Dimethyldioxirane (DMDO). Org. Synth. 2013, 90, 350–357. [Google Scholar]







© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Lin, D.; Guo, X.; Huang, W.; Zheng, J.; Peng, Y. Chemical Identity of Interaction of Protein with Reactive Metabolite of Diosbulbin B In Vitro and In Vivo. Toxins 2017, 9, 249. https://doi.org/10.3390/toxins9080249
Wang K, Lin D, Guo X, Huang W, Zheng J, Peng Y. Chemical Identity of Interaction of Protein with Reactive Metabolite of Diosbulbin B In Vitro and In Vivo. Toxins. 2017; 9(8):249. https://doi.org/10.3390/toxins9080249
Chicago/Turabian StyleWang, Kai, Dongju Lin, Xiucai Guo, Wenlin Huang, Jiang Zheng, and Ying Peng. 2017. "Chemical Identity of Interaction of Protein with Reactive Metabolite of Diosbulbin B In Vitro and In Vivo" Toxins 9, no. 8: 249. https://doi.org/10.3390/toxins9080249
APA StyleWang, K., Lin, D., Guo, X., Huang, W., Zheng, J., & Peng, Y. (2017). Chemical Identity of Interaction of Protein with Reactive Metabolite of Diosbulbin B In Vitro and In Vivo. Toxins, 9(8), 249. https://doi.org/10.3390/toxins9080249