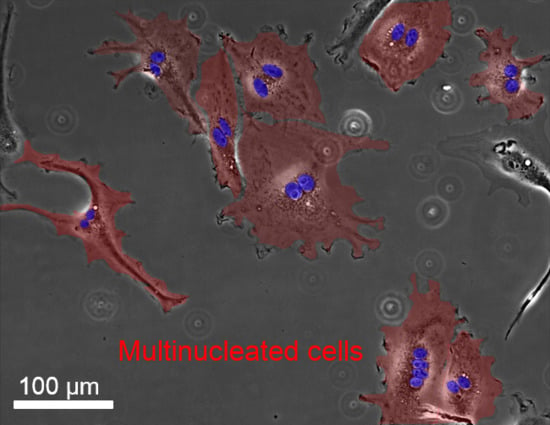
Multinucleation of Incubated Cells and Their Morphological Differences Compared to Mononuclear Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Contact Angle of Dishes
2.2. Cells
2.3. Conditions Used to Prepare Multinucleated Cells
2.4. Time-Lapse Imaging
2.5. Morphometry of the Cells and Cell Nuclei
2.6. Statistical Analysis
3. Results and Discussion
3.1. Contact Angle of Glass Bottom Dishes
3.2. Comparison of Multinucleated Cells Plated on Different Dishes
3.3. Changes in the Number of Nuclei during the Incubation
3.4. Morphology of the Cells and Cell Nuclei of Multinucleated and Mononuclear Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Alberts, B.; Wilson, J.H.; Johnson, A.; Hunt, T.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell: Reference edition, 5th ed.; John, H., Wilson, T.H., Eds.; Garland Science: New York, NY, USA, 2008; p. 1099. [Google Scholar]
- Helming, L.; Gordon, S. Macrophage fusion induced by IL-4 alternative activation is a multistage process involving multiple target molecules. Eur. J. Immunol. 2007, 37, 33–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedlak, B.J.; Booyse, F.M.; Bell, S.; Rafelson Jr., M.E. Comparison of two types of endothelial cells in long term culture. Thromb. Haemost. 1976, 35, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, O.; Fan, J.L.; Watanabe, T. Atherosclerosis- and age-related multinucleated variant endothelial cells in primary culture from human aorta. Am. J. Pathol. 1989, 135, 967–976. [Google Scholar] [PubMed]
- Ariizumi, T.; Ogose, A.; Kawashima, H.; Hotta, T.; Umezu, H.; Endo, N. Multinucleation followed by an acytokinetic cell division in myxofibrosarcoma with giant cell proliferation. J. Exp. Clin. Cancer Res. 2009, 28, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boșca, A.B.; Ilea, A.; Eovrea, A.S.; Constantin, A.M.; Ruxanda, F.; Rus, V.; Rațiu, C.; Miclăuș, V. Multinucleated Giant Cells Polymorphism in Epulis. Bull. Univ. Agric. Sci. Vet. Med. Cluj.-Napoca. Agric. 2015, 72, 47–52. [Google Scholar] [CrossRef]
- Herring, M.; Gardner, A.; Glover, J. A single-staged technique for seeding vascular grafts with autogenous endothelium. Surgery 1978, 84, 498–504. [Google Scholar] [PubMed]
- Zang, J.H.; Cavet, G.; Sabry, J.H.; Wagner, P.; Moores, S.L.; Spudich, J.A. On the role of myosin-II in cytokinesis: division of Dictyostelium cells under adhesive and nonadhesive conditions. Mol. Biol. Cell 1997, 8, 2617–2629. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.; Witkin, K.L.; Cohen-Fix, O. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 2009, 122, 1477–1486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zink, D.; Fischer, A.H.; Nickerson, J.A. Nuclear structure in cancer cells. Nat. Rev. Cancer 2004, 4, 677. [Google Scholar] [CrossRef] [PubMed]
- Kohli, R.; Mittal, K.L. Developments in Surface Contamination and Cleaning—Vol 6: Methods of Cleaning and Cleanliness Verification, 1st ed.; Elsevier Science: Waltham, MA, USA, 2013; p. 169. [Google Scholar]
- Shi, Q.; King, R.W. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature 2005, 437, 1038. [Google Scholar] [CrossRef] [PubMed]
- Draviam, V.M.; Xie, S.; Sorger, P.K. Chromosome segregation and genomic stability. Curr. Opin. Genet. Dev. 2004, 14, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, S.; Amon, A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 2015, 16, 473. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.L.; Heald, R. Nuclear Size Is Regulated by Importin α and Ntf2 in Xenopus. Cell 2010, 143, 288–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camps, J.; Erdos, M.R.; Ried, T. The role of lamin B1 for the maintenance of nuclear structure and function. Nucleus 2015, 6, 8–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, R.; Nieto, S.; Santamaria, L. Stereologic estimates of volume-weighted mean nuclear volume in colorectal adenocarcinoma: correlation with histologic grading, Dukes’ staging, cell proliferation activity and p53 protein expression. Gen. Diagn. Pathol. 1997, 143, 29–38. [Google Scholar] [PubMed]
- Jackson, B.; Peyrollier, K.; Pedersen, E.; Basse, A.; Karlsson, R.; Wang, Z.; Lefever, T.; Ochsenbein, A.M.; Schmidt, G.; Aktories, K.; et al. RhoA is dispensable for skin development, but crucial for contraction and directed migration of keratinocytes. Mol. Biol. Cell 2011, 22, 593–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, E.B. The Cell in Development and Inheritance, 3rd ed.; Macmillan Company: New York, NY, USA, 1925; pp. 727–739. [Google Scholar]
- Yamamoto, T.; Horiguchi, H.; Kamma, H.; Ogata, T.; Fukasawa, M.; Ikezawa, T.; Inage, Y.; Akaogi, E.; Mitsui, K.; Hori, M. The effect of nuclear DNA content on nuclear atypia and clinicopathological factors in non-small cell lung carcinoma. J. Jpn. Soc. Clin. Cytol. 1993, 32, 846–852. [Google Scholar] [CrossRef]
- Omelchenko, T.; Vasiliev, J.M.; Gelfand, I.M.; Feder, H.H.; Bonder, E.M. Mechanisms of polarization of the shape of fibroblasts and epitheliocytes: Separation of the roles of microtubules and Rho-dependent actin-myosin contractility. Proc. Natl. Acad. Sci. USA 2002, 99, 10452–10457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babich, A.; Li, S.; O’Connor, R.S.; Milone, M.C.; Freedman, B.D.; Burkhardt, J.K. F-actin polymerization and retrograde flow drive sustained PLCγ1 signaling during T cell activation. J. Cell Biol. 2012, 197, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Kharitonova, M.A.; Vasiliev, J.M. Length control is determined by the pattern of cytoskeleton. J. Cell Sci. 2004, 117, 1955–1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, B.C.; Kelley, L.C.; Loskutov, Y.V.; Marinak, K.M.; Kozyreva, V.K.; Smolkin, M.B.; Pugacheva, E.N. Dual Targeting of Mesenchymal and Amoeboid Motility Hinders Metastatic Behavior. Mol. Cancer Res. 2017, 15, 670–682. [Google Scholar] [CrossRef] [PubMed]
- Kazmers, N.H.; Ma, S.A.; Yoshida, T.; Stern, P.H. Rho GTPase signaling and PTH 3–34, but not PTH 1–34, maintain the actin cytoskeleton and antagonize bisphosphonate effects in mouse osteoblastic MC3T3-E1 cells. Bone 2009, 45, 52–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sugita, S.; Munechika, R.; Nakamura, M. Multinucleation of Incubated Cells and Their Morphological Differences Compared to Mononuclear Cells. Micromachines 2019, 10, 156. https://doi.org/10.3390/mi10020156
Sugita S, Munechika R, Nakamura M. Multinucleation of Incubated Cells and Their Morphological Differences Compared to Mononuclear Cells. Micromachines. 2019; 10(2):156. https://doi.org/10.3390/mi10020156
Chicago/Turabian StyleSugita, Shukei, Risa Munechika, and Masanori Nakamura. 2019. "Multinucleation of Incubated Cells and Their Morphological Differences Compared to Mononuclear Cells" Micromachines 10, no. 2: 156. https://doi.org/10.3390/mi10020156
APA StyleSugita, S., Munechika, R., & Nakamura, M. (2019). Multinucleation of Incubated Cells and Their Morphological Differences Compared to Mononuclear Cells. Micromachines, 10(2), 156. https://doi.org/10.3390/mi10020156