Prognostic Implication of pAMPK Immunohistochemical Staining by Subcellular Location and Its Association with SMAD Protein Expression in Clear Cell Renal Cell Carcinoma
Abstract
:1. Introduction
2. Results
2.1. Patients and pAMPK IHC Staining
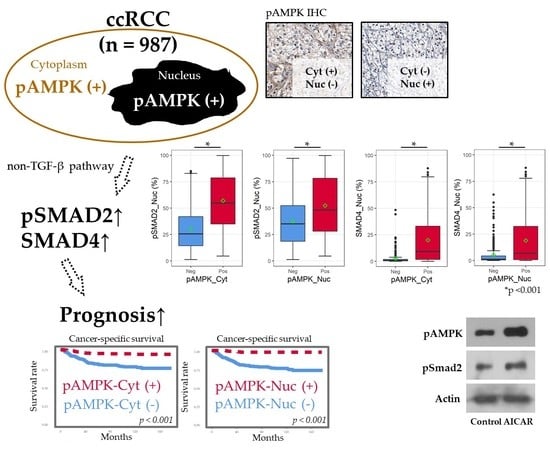
2.2. Positive IHC Staining for pAMPK Was Significantly Associated with Improved ccRCC Prognosis
2.3. pAMPK Induced Nuclear SMAD Protein Expression in ccRCC
3. Discussion
4. Materials and Methods
4.1. Patients’ Cohorts
4.2. TMA Construction and IHC Staining
4.3. Establishment of Cut-Off Criteria for pAMPK IHC Staining Positivity
4.4. Western Blot Analysis
4.5. Characteristics of the TCGA ccRCC Dataset
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massari, F.; Ciccarese, C.; Santoni, M.; Brunelli, M.; Piva, F.; Modena, A.; Bimbatti, D.; Fantinel, E.; Santini, D.; Cheng, L.; et al. Metabolic alterations in renal cell carcinoma. Cancer Treat. Rev. 2015, 41, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, P.T.; Hay, N. The two TORCs and Akt. Dev. Cell 2007, 12, 487–502. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Jung, M.; Moon, K.C. The prognostic significance of nuclear expression of PHF2 and C/EBPalpha in clear cell renal cell carcinoma with consideration of adipogenic metabolic evolution. Oncotarget 2018, 9, 142–151. [Google Scholar] [CrossRef]
- Jung, M.; Lee, C.; Park, J.H.; Moon, K.C. Prognostic significance of immunohistochemical staining for myoferlin in clear cell renal cell carcinoma and its association with epidermal growth factor receptor expression. Urol. Oncol. 2019, in press. [Google Scholar] [CrossRef]
- Ricketts, C.J.; De Cubas, A.A.; Fan, H.; Smith, C.C.; Lang, M.; Reznik, E.; Bowlby, R.; Gibb, E.A.; Akbani, R.; Beroukhim, R.; et al. The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. 2018, 23, 313–326.e5. [Google Scholar] [CrossRef]
- Shackelford, D.B.; Shaw, R.J. The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat. Rev. Cancer 2009, 9, 563–575. [Google Scholar] [CrossRef]
- Tamargo-Gomez, I.; Marino, G. AMPK: Regulation of metabolic dynamics in the context of autophagy. Int. J. Mol. Sci. 2018, 19, 3812. [Google Scholar] [CrossRef]
- Woodard, J.; Joshi, S.; Viollet, B.; Hay, N.; Platanias, L.C. AMPK as a therapeutic target in renal cell carcinoma. Cancer Biol. Ther. 2010, 10, 1169–1178. [Google Scholar] [CrossRef]
- Choi, C.H.; Chung, J.Y.; Cho, H.; Kitano, H.; Chang, E.; Ylaya, K.; Chung, E.J.; Kim, J.H.; Hewitt, S.M. Prognostic significance of AMP-dependent kinase alpha expression in cervical cancer. Pathobiology 2015, 82, 203–211. [Google Scholar] [CrossRef]
- Li, C.L.; Liu, V.W.S.; Chiu, P.M.; Chan, D.W.; Ngan, H.Y.S. Over-expressions of AMPK subunits in ovarian carcinomas with significant clinical implications. BMC Cancer 2012, 12, 357. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Yang, W.; Wu, F.; Wang, C.; Yu, L.; Tang, L.; Qiu, B.; Li, Y.; Guo, L.; Wu, M.; et al. Prognostic significance of AMPK activation and therapeutic effects of metformin in hepatocellular carcinoma. Clin. Cancer Res. 2013, 19, 5372–5380. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.C.; Chien, M.H.; Liu, H.Y.; Chang, Y.C.; Chen, C.K.; Lee, W.J.; Kuo, T.C.; Hsiao, M.; Hua, K.T.; Cheng, T.Y. Nuclear translocation of PKM2/AMPK complex sustains cancer stem cell populations under glucose restriction stress. Cancer Lett. 2018, 421, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Z.; Wang, H.; Chen, X.; Jin, C. Activation of AMPK by metformin promotes renal cancer cell proliferation under glucose deprivation through its interaction with PKM2. Int. J. Biol. Sci. 2019, 15, 617–627. [Google Scholar] [CrossRef]
- Mishra, R.; Cool, B.L.; Laderoute, K.R.; Foretz, M.; Viollet, B.; Simonson, M.S. AMP-activated protein kinase inhibits transforming growth factor-beta-induced Smad3-dependent transcription and myofibroblast transdifferentiation. J. Biol. Chem. 2008, 283, 10461–10469. [Google Scholar] [CrossRef]
- Lim, J.Y.; Oh, M.A.; Kim, W.H.; Sohn, H.Y.; Park, S.I. AMP-activated protein kinase inhibits TGF-beta-induced fibrogenic responses of hepatic stellate cells by targeting transcriptional coactivator p300. J. Cell. Physiol. 2012, 227, 1081–1089. [Google Scholar] [CrossRef]
- Stone, J.D.; Holt, A.W.; Vuncannon, J.R.; Brault, J.J.; Tulis, D.A. AMP-activated protein kinase inhibits transforming growth factor-beta-mediated vascular smooth muscle cell growth: Implications for a Smad-3-dependent mechanism. Am. J. Physiol. Heart Circ. Physiol. 2015, 309, H1251–H1259. [Google Scholar] [CrossRef]
- Zhao, J.; Miyamoto, S.; You, Y.H.; Sharma, K. AMP-activated protein kinase (AMPK) activation inhibits nuclear translocation of Smad4 in mesangial cells and diabetic kidneys. Am. J. Physiol. Ren. Physiol. 2015, 308, F1167–F1177. [Google Scholar] [CrossRef] [Green Version]
- Yadav, H.; Devalaraja, S.; Chung, S.T.; Rane, S.G. TGF-1/Smad3 pathway targets PP2A-AMPK-FoxO1 signaling to regulate hepatic gluconeogenesis. J. Biol. Chem. 2017, 292, 3420–3432. [Google Scholar] [CrossRef]
- Li, N.S.; Zou, J.R.; Lin, H.; Ke, R.; He, X.L.; Xiao, L.; Huang, D.; Luo, L.; Lv, N.; Luo, Z. LKB1/AMPK inhibits TGF-beta1 production and the TGF-beta signaling pathway in breast cancer cells. Tumour Biol. 2016, 37, 8249–8258. [Google Scholar] [CrossRef]
- Tang, J.; Gifford, C.C.; Samarakoon, R.; Higgins, P.J. Deregulation of negative controls on TGF-beta1 signaling in tumor progression. Cancers 2018, 10, 159. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Lee, C.; Suh, J.H.; Chae, J.Y.; Moon, K.C. Nuclear expression of Smad proteins and its prognostic significance in clear cell renal cell carcinoma. Hum. Pathol. 2013, 44, 2047–2054. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.C.; Lee, K.H.; Lai, I.L.; Wang, D.; Mo, X.; Kulp, S.K.; Shapiro, C.L.; Chen, C.S. AMPK reverses the mesenchymal phenotype of cancer cells by targeting the Akt-MDM2-Foxo3a signaling axis. Cancer Res. 2014, 74, 4783–4795. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Huang, D.; Lu, N.; Luo, L. Role of the LKB1/AMPK pathway in tumor invasion and metastasis of cancer cells (Review). Oncol. Rep. 2015, 34, 2821–2826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, S.; Medina, E.A.; Li, B.; Habib, S.L. Preclinical evidence of the enhanced effectiveness of combined rapamycin and AICAR in reducing kidney cancer. Mol. Oncol. 2018, 12, 1917–1934. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Fan, Y.; Huang, S.; Wang, G.; Han, R.; Lei, F.; Luo, A.; Jing, X.; Zhao, L.; Gu, S.; et al. Thymoquinone inhibits the metastasis of renal cell cancer cells by inducing autophagy via AMPK/mTOR signaling pathway. Cancer Sci. 2018, 109, 3865–3873. [Google Scholar] [CrossRef]
- Quentin, T.; Kitz, J.; Steinmetz, M.; Poppe, A.; Bar, K.; Kratzner, R. Different expression of the catalytic alpha subunits of the AMP activated protein kinase--an immunohistochemical study in human tissue. Histol. Histopathol. 2011, 26, 589–596. [Google Scholar] [CrossRef]
- Vidal, A.P.; Andrade, B.M.; Vaisman, F.; Cazarin, J.; Pinto, L.F.R.; Breitenbach, M.M.D.; Corbo, R.; Caroli-Bottino, A.; Soares, F.; Vaisman, M.; et al. AMP-activated protein kinase signaling is upregulated in papillary thyroid cancer. Eur. J. Endocrinol. 2013, 169, 521–528. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, K. Smad phosphoisoform signaling specificity: The right place at the right time. Carcinogenesis 2011, 32, 1578–1588. [Google Scholar] [CrossRef]
- Katajisto, P.; Vaahtomeri, K.; Ekman, N.; Ventela, E.; Ristimaki, A.; Bardeesy, N.; Feil, R.; DePinho, R.A.; Makela, T.P. LKB1 signaling in mesenchymal cells required for suppression of gastrointestinal polyposis. Nat. Genet. 2008, 40, 455–459. [Google Scholar] [CrossRef]
- Vaahtomeri, K.; Ventela, E.; Laajanen, K.; Katajisto, P.; Wipff, P.J.; Hinz, B.; Vallenius, T.; Tiainen, M.; Makela, T.P. Lkb1 is required for TGFbeta-mediated myofibroblast differentiation. J. Cell Sci. 2008, 121, 3531–3540. [Google Scholar] [CrossRef]
- Fu, G.; Peng, C. Nodal enhances the activity of FoxO3a and its synergistic interaction with Smads to regulate cyclin G2 transcription in ovarian cancer cells. Oncogene 2011, 30, 3953–3966. [Google Scholar] [CrossRef] [Green Version]
- Gomis, R.R.; Alarcon, C.; He, W.; Wang, Q.; Seoane, J.; Lash, A.; Massague, J. A FoxO-smad synexpression group in human keratinocytes. Proc. Natl. Acad. Sci. USA 2006, 103, 12747–12752. [Google Scholar] [CrossRef]
- Sridharan, S.; Jain, K.; Basu, A. Regulation of autophagy by kinases. Cancers 2011, 3, 2630–2654. [Google Scholar] [CrossRef]
- Paner, G.P.; Stadler, W.M.; Hansel, D.E.; Montironi, R.; Lin, D.W.; Amin, M.B. Updates in the eighth edition of the tumor-node-metastasis staging classification for urologic cancers. Eur. Urol. 2018, 73, 560–569. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Broad Institute GDAC Firehose. Available online: http://gdac.broadinstitute.org/ (accessed on 5 August 2019).
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]




| Discovery Cohort | Cyt Pos 1 | Cyt Neg | p | Nuc Pos 1 | Nuc Neg | p | Total |
|---|---|---|---|---|---|---|---|
| Number | n = 250 (55.2%) | n = 203 (44.8%) | n = 228 (50.3%) | n = 225 (49.7%) | 453 | ||
| Age (year) | 0.006 | 0.009 | |||||
| ≥58 | 118 (47.2%) | 123 (60.6%) | 107 (46.9%) | 134 (59.6%) | 241 (53.2%) | ||
| <58 | 132 (52.8%) | 80 (39.4%) | 121 (53.1%) | 91 (40.4%) | 212 (46.8%) | ||
| Sex | 0.483 | 0.003 | |||||
| Male | 180 (72.0%) | 153 (75.4%) | 153 (67.1%) | 180 (80.0%) | 333 (73.5%) | ||
| Female | 70 (28.0%) | 50 (24.6%) | 75 (32.9%) | 45 (20.0%) | 120 (26.5%) | ||
| Size (cm)2 | 3.0 [2.0–4.5] | 5.0 [3.0–7.9] | <0.001 3 | 3.0 [2.0–4.7] | 4.5 [3.0–7.5] | <0.001 3 | 3.5 [2.3–6.0] |
| TNM stage | <0.001 | <0.001 | |||||
| Low (I or II) | 219 (87.6%) | 138 (68.0%) | 209 (91.7%) | 148 (65.8%) | 357 (78.8%) | ||
| High (III or IV) | 31 (12.4%) | 65 (32.0%) | 19 (8.3%) | 77 (34.2%) | 96 (21.2%) | ||
| WHO grade | <0.001 | <0.001 | |||||
| Low (1 or 2) | 141 (56.4%) | 79 (38.9%) | 155 (68.0%) | 65 (28.9%) | 220 (48.6%) | ||
| High (3 or 4) | 109 (43.6%) | 124 (61.1%) | 73 (32.0%) | 160 (71.1%) | 233 (51.4%) | ||
| Validation cohort | Cyt Pos 1 | Cyt Neg | p | Nuc Pos 1 | Nuc Neg | p | Total |
| Number | n = 242 (45.3%) | n = 292 (54.7%) | n = 231 (43.3%) | n = 303 (56.7%) | 534 | ||
| Age (year) | 0.078 | 0.045 | |||||
| ≥56 | 114 (47.1%) | 161 (55.1%) | 107 (46.3%) | 168 (55.4%) | 275 (51.5%) | ||
| <56 | 128 (52.9%) | 131 (44.9%) | 124 (53.7%) | 135 (44.6%) | 259 (48.5%) | ||
| Sex | 0.737 | 0.001 | |||||
| Male | 183 (75.6%) | 216 (74.0%) | 156 (67.5%) | 243 (80.2%) | 399 (74.7%) | ||
| Female | 59 (24.5%) | 76 (26.0%) | 75 (32.5%) | 60 (19.8%) | 135 (25.3%) | ||
| Size (cm) 2 | 4.0 [3.0–6.0] | 5.5 [3.8–8.8] | <0.001 3 | 4.0 [3.0–6.5] | 5.3 [3.9–8.0] | <0.001 3 | 4.8 [3.2–7.5] |
| TNM stage | <0.001 | <0.001 | |||||
| Low (I or II) | 204 (84.3%) | 180 (61.6%) | 191 (82.7%) | 193 (63.7%) | 384 (71.9%) | ||
| High (III or IV) | 38 (15.7%) | 112 (38.4%) | 40 (17.3%) | 110 (36.3%) | 150 (28.1%) | ||
| WHO grade | 0.085 | <0.001 | |||||
| Low (1 or 2) | 140 (57.9%) | 146 (50.0%) | 155 (67.1%) | 131 (43.2%) | 286 (53.6%) | ||
| High (3 or 4) | 102 (42.1%) | 146 (50.0%) | 76 (32.9%) | 172 (56.8%) | 248 (46.4%) |
| Analysis Detail | Progression-Free Survival | Overall Survival | Cancer-Specific Survival | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | |
| Univariate analysis | ||||||
| pAMPK-C (Pos vs Neg) | 0.190 (0.110–0.310) | <0.001 | 0.470 (0.320–0.680) | <0.001 | 0.200 (0.110–0.370) | <0.001 |
| pAMPK-N (Pos vs Neg) | 0.140 (0.080–0.260) | <0.001 | 0.440 (0.300–0.650) | <0.001 | 0.070 (0.030–0.19) | <0.001 |
| TNM stage (≥III vs ≤II) | 12.920 (8.150–20.490) | <0.001 | 5.480 (3.790–7.920) | <0.001 | 18.050 (10.100–32.270) | <0.001 |
| WHO Grade (≥3 vs ≤2) | 5.210 (2.980–9.120) | <0.001 | 2.770 (1.850–4.160) | <0.001 | 16.330 (5.930–44.950) | <0.001 |
| Multivariate analysis | ||||||
| pAMPK-C (Pos vs Neg) | 0.260 (0.153–0.442) | <0.001 | 0.656 (0.446–0.965) | 0.032 | 0.374 (0.205–0.681) | 0.001 |
| TNM stage (≥III vs ≤II) | 8.644 (5.340–13.992) | <0.001 | 4.163 (2.806–6.178) | <0.001 | 9.535 (5.245–17.336) | <0.001 |
| WHO Grade (≥3 vs ≤2) | 2.601 (1.456–4.646) | 0.001 | 1.774 (1.156–2.724) | 0.009 | 7.163 (2.552–20.106) | <0.001 |
| Multivariate analysis | ||||||
| pAMPK-N (Pos vs Neg) | 0.308 (0.159–0.595) | <0.001 | 0.767 (0.500–1.177) | 0.225 | 0.232 (0.090–0.600) | 0.003 |
| TNM stage (≥III vs ≤II) | 7.944 (4.868–12.965) | <0.001 | 4.250 (2.850–6.337) | <0.001 | 8.677 (4.754–15.837) | <0.001 |
| WHO Grade (≥3 vs ≤2) | 1.889 (1.024–3.487) | 0.042 | 1.696 (1.082–2.660) | 0.021 | 5.086 (1.777–14.556) | 0.002 |
| Analysis Detail | Progression-Free Survival | Overall Survival | Cancer-Specific Survival | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | |
| Univariate analysis | ||||||
| pAMPK–C (Pos vs Neg) | 0.250 (0.180–0.360) | <0.001 | 0.480 (0.340–0.690) | <0.001 | 0.180 (0.100–0.310) | <0.001 |
| pAMPK–N (Pos vs Neg) | 0.300 (0.210–0.440) | <0.001 | 0.350 (0.230–0.510) | <0.001 | 0.180 (0.100–0.330) | <0.001 |
| TNM stage (≥III vs ≤II) | 6.430 (4.720–8.760) | <0.001 | 4.740 (3.390–6.620) | <0.001 | 10.340 (6.570–16.270) | <0.001 |
| WHO Grade (≥3 vs ≤2) | 3.010 (2.190–4.140) | <0.001 | 2.870 (2.020–4.080) | <0.001 | 4.640 (2.880–7.470) | <0.001 |
| Multivariate analysis | ||||||
| pAMPK–C (Pos vs Neg) | 0.304 (0.210–0.441) | <0.001 | 0.629 (0.438–0.903) | 0.012 | 0.256 (0.144–0.455) | <0.001 |
| TNM stage (≥III vs ≤II) | 4.630 (3.352–6.395) | <0.001 | 3.567 (2.505–5.079) | <0.001 | 6.446 (4.023–10.328) | <0.001 |
| WHO Grade (≥3 vs ≤2) | 2.244 (1.618–3.112) | <0.001 | 2.091 (1.453–3.010) | <0.001 | 2.935 (1.801–4.781) | <0.001 |
| Multivariate analysis | ||||||
| pAMPK–N (Pos vs Neg) | 0.405 (0.280–0.585) | <0.001 | 0.471 (0.315–0.705) | <0.001 | 0.296 (0.164–0.536) | <0.001 |
| TNM stage (≥III vs ≤II) | 4.989 (3.617–6.883) | <0.001 | 3.601 (2.537–5.111) | <0.001 | 7.101 (4.434–11.371) | <0.001 |
| WHO Grade (≥3 vs ≤2) | 1.882 (1.350–2.623) | <0.001 | 1.844 (1.274–2.667) | 0.001 | 2.344 (1.430–3.844) | <0.001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, M.; Lee, J.H.; Lee, C.; Park, J.H.; Park, Y.R.; Moon, K.C. Prognostic Implication of pAMPK Immunohistochemical Staining by Subcellular Location and Its Association with SMAD Protein Expression in Clear Cell Renal Cell Carcinoma. Cancers 2019, 11, 1602. https://doi.org/10.3390/cancers11101602
Jung M, Lee JH, Lee C, Park JH, Park YR, Moon KC. Prognostic Implication of pAMPK Immunohistochemical Staining by Subcellular Location and Its Association with SMAD Protein Expression in Clear Cell Renal Cell Carcinoma. Cancers. 2019; 11(10):1602. https://doi.org/10.3390/cancers11101602
Chicago/Turabian StyleJung, Minsun, Jeong Hoon Lee, Cheol Lee, Jeong Hwan Park, Yu Rang Park, and Kyung Chul Moon. 2019. "Prognostic Implication of pAMPK Immunohistochemical Staining by Subcellular Location and Its Association with SMAD Protein Expression in Clear Cell Renal Cell Carcinoma" Cancers 11, no. 10: 1602. https://doi.org/10.3390/cancers11101602
APA StyleJung, M., Lee, J. H., Lee, C., Park, J. H., Park, Y. R., & Moon, K. C. (2019). Prognostic Implication of pAMPK Immunohistochemical Staining by Subcellular Location and Its Association with SMAD Protein Expression in Clear Cell Renal Cell Carcinoma. Cancers, 11(10), 1602. https://doi.org/10.3390/cancers11101602