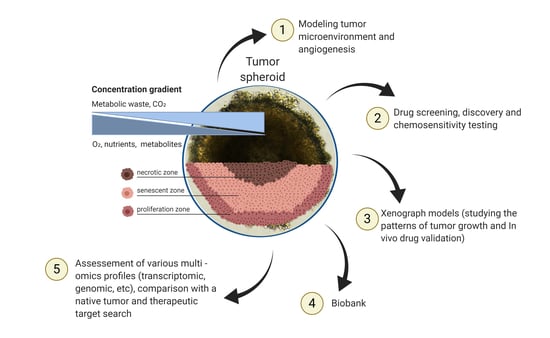
Promising Applications of Tumor Spheroids and Organoids for Personalized Medicine
Abstract
:Simple Summary
Abstract
1. Introduction
2. Tumor Spheroids and Organoids
3. Methods for Obtaining Spheroids and Organoids
| 3D Models | Materials for 3D Models Preparation | Advantages | Disadvantages |
|---|---|---|---|
| Spheroids [19,29,55,67,68,69,70,71] | (1) Various plates and rotors can be used for cultivation (spinner flasks, rotary cell culture systems, poly-2-hydroxyethyl methacrylate (poly-Hema)-coated plates, liquid overlay, micropatterned plates, low binding plates, microfluidics device). (2) The addition of B27, epidermal growth factor (EGF) and fibroblast growth factors (FGFs) are essential for spheres obtained from CSCs. Spheroids can be cultivated, including fetal bovine serum (FBS) and without it. | (1) Possibility of: (a) Creation of co-culture. (b) Culturing cells without special equipment. (c) Cultures without expensive cultivation methods. (2) Gas, nutrient and pH gradients are present. | (1) The difficulty of forming homogeneous spheroids. (2) Fragile spheroid structure. (3) Gradient structure complicates drug testing. |
| Organoids [44,72,73,74,75] | (1) Cells and cell aggregates culturing are cultured on various matrices (Matrigel, collagen type I, HA (hyaluronic acid) hydrogel, PEG hydrogel, Fibrin/laminin hydrogel). (2) Culture supplements depending on the tissue type are essential. The main ones include: Wnt, nicotinamid, N-acetylcysteine, R-spondin-1, FGFs, noggin and molecule inhibitors (Y27632, A-83-01, SB202190). | (1) Possibility of: (a) Creation of co-culture. (b) Reproduction of intercellular interactions and cell-ECM interactions. (c) Primary tumor cells long-term cultivation. (2) Stable at passaging. | (1) Expensive method. (2) The gradient of gases, nutrients and pH is not always reproducible. (3) Therapeutic responses may depend on the matrix. |
4. Spheroids and Organoids for Personalized Medicine
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Krzyszczyk, P.; Acevedo, A.; Davidoff, E.J.; Timmins, L.M.; Marrero-Berrios, I.; Patel, M.; White, C.; Lowe, C.; Sherba, J.J.; Hartmanshenn, C.; et al. The growing role of precision and personalized medicine for cancer treatment. Technology 2018, 6, 79–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, M. Personalized medicine and cancer. J. Pers. Med. 2012, 2, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reynies, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef] [PubMed]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef] [Green Version]
- Rivenbark, A.G.; O’Connor, S.M.; Coleman, W.B. Molecular and cellular heterogeneity in breast cancer: Challenges for personalized medicine. Am. J. Pathol. 2013, 183, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Offit, K. Personalized medicine: New genomics, old lessons. Hum. Genet. 2011, 130, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Kitaeva, K.V.; Rutland, C.S.; Rizvanov, A.A.; Solovyeva, V.V. Cell culture Based in vitro test systems for anticancer drug screening. Front. Bioeng. Biotechnol. 2020, 8, 322. [Google Scholar] [CrossRef] [Green Version]
- Mingaleeva, R.N.; Solovieva, V.V.; Blatt, N.L.; Rizvanov, A.A. Application of cell and tissue cultures for potential anti-cancer/oncology drugs screening in vitro. Cell. Transplant. Tissue Eng. 2013, 8, 20–28. [Google Scholar]
- Kitaeva, K.V.; Prudnikov, T.S.; Gomzikova, M.O.; Kletukhina, S.K.; James, V.; Rizvanov, A.A.; Solovyeva, V.V. Analysis of the interaction and proliferative activity of adenocarcinoma, peripheral blood mononuclear and mesenchymal stromal cells after co-cultivation in vitro. BioNanoScience 2019, 9, 502–509. [Google Scholar] [CrossRef]
- Ham, S.L.; Joshi, R.; Thakuri, P.S.; Tavana, H. Liquid-based three-dimensional tumor models for cancer research and drug discovery. Exp. Biol. Med. 2016, 241, 939–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Su, J.; Fu, X.; Zheng, L.; Yin, Z. Microfluidic device for primary tumor spheroid isolation. Exp. Hematol. Oncol. 2017, 6, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chulpanova, D.S.; Kitaeva, K.V.; Rutland, C.S.; Rizvanov, A.A.; Solovyeva, V.V. Mouse tumor models for advanced cancer immunotherapy. Int. J. Mol. Sci. 2020, 21, 4118. [Google Scholar] [CrossRef] [PubMed]
- Perone, Y.; Farrugia, A.J.; Meira, A.R.; Gyorffy, B.; Ion, C.; Uggetti, A.; Chronopoulos, A.; Marrazzo, P.; Faronato, M.; Shousha, S.; et al. SREBP1 drives Keratin-80-dependent cytoskeletal changes and invasive behavior in endocrine-resistant ER alpha breast cancer. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Capp, J.P. Cancer stem cells: From historical roots to a new perspective. J. Oncol. 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Konishi, J.; Kawaguchi, K.S.; Vo, H.; Haruki, N.; Gonzalez, A.; Carbone, D.P.; Dang, T.P. gamma-secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 2007, 67, 8051–8057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hynds, R.E.; Giangreco, A. Concise review: The relevance of human stem cell-derived organoid models for epithelial translational medicine. Stem Cells 2013, 31, 417–422. [Google Scholar] [CrossRef] [Green Version]
- Weiswald, L.B.; Bellet, D.; Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Eglen, R.M. Three-dimensional cell cultures in drug discovery and development. SLAS Discov. Adv. Life Sci. RD 2017, 22, 456–472. [Google Scholar] [CrossRef] [Green Version]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef] [Green Version]
- Ekert, J.E.; Johnson, K.; Strake, B.; Pardinas, J.; Jarantow, S.; Perkinson, R.; Colter, D.C. Three-dimensional lung tumor microenvironment modulates therapeutic compound responsiveness in vitro—Implication for drug development. PLoS ONE 2014, 9, e92248. [Google Scholar] [CrossRef]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H. The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Hirschhaeuser, F.; Menne, H.; Dittfeld, C.; West, J.; Mueller-Klieser, W.; Kunz-Schughart, L.A. Multicellular tumor spheroids: An underestimated tool is catching up again. J. Biotechnol. 2010, 148, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Alfarouk, K.O.; Verduzco, D.; Rauch, C.; Muddathir, A.K.; Adil, H.H.; Elhassan, G.O.; Ibrahim, M.E.; David Polo Orozco, J.; Cardone, R.A.; Reshkin, S.J.; et al. Glycolysis, tumor metabolism, cancer growth and dissemination. A new pH-based etiopathogenic perspective and therapeutic approach to an old cancer question. Oncoscience 2014, 1, 777–802. [Google Scholar] [CrossRef]
- Martin, A.R.; Ronco, C.; Demange, L.; Benhida, R. Hypoxia inducible factor down-regulation, cancer and cancer stem cells (CSCs): Ongoing success stories. MedChemComm 2017, 8, 21–52. [Google Scholar] [CrossRef]
- Gupta, P.B.; Onder, T.T.; Jiang, G.; Tao, K.; Kuperwasser, C.; Weinberg, R.A.; Lander, E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 2009, 138, 645–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oskarsson, T.; Acharyya, S.; Zhang, X.H.; Vanharanta, S.; Tavazoie, S.F.; Morris, P.G.; Downey, R.J.; Manova-Todorova, K.; Brogi, E.; Massague, J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat. Med. 2011, 17, 867–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maliszewska-Olejniczak, K.; Brodaczewska, K.K.; Bielecka, Z.F.; Solarek, W.; Kornakiewicz, A.; Szczylik, C.; Porta, C.; Czarnecka, A.M. Development of extracellular matrix supported 3D culture of renal cancer cells and renal cancer stem cells. Cytotechnology 2019, 71, 149–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishiguro, T.; Ohata, H.; Sato, A.; Yamawaki, K.; Enomoto, T.; Okamoto, K. Tumor-derived spheroids: Relevance to cancer stem cells and clinical applications. Cancer Sci. 2017, 108, 283–289. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Ponti, D.; Costa, A.; Zaffaroni, N.; Pratesi, G.; Petrangolini, G.; Coradini, D.; Pilotti, S.; Pierotti, M.A.; Daidone, M.G. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005, 65, 5506–5511. [Google Scholar] [CrossRef] [Green Version]
- Todaro, M.; Alea, M.P.; Di Stefano, A.B.; Cammareri, P.; Vermeulen, L.; Iovino, F.; Tripodo, C.; Russo, A.; Gulotta, G.; Medema, J.P.; et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 2007, 1, 389–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallas, N.A.; Xia, L.; Fan, F.; Gray, M.J.; Gaur, P.; van Buren, G.; Samuel, S.; Kim, M.P.; Lim, S.J.; Ellis, L.M. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009, 69, 1951–1957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, J.; Endo, H.; Okuyama, H.; Ishikawa, O.; Iishi, H.; Tsujii, M.; Ohue, M.; Inoue, M. Retaining cell-cell contact enables preparation and culture of spheroids composed of pure primary cancer cells from colorectal cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 6235–6240. [Google Scholar] [CrossRef] [Green Version]
- Ishiguro, T.; Sato, A.; Ohata, H.; Ikarashi, Y.; Takahashi, R.; Ochiya, T.; Yoshida, M.; Tsuda, H.; Onda, T.; Kato, T.; et al. Establishment and characterization of an in vitro model of ovarian cancer stem-like cells with an enhanced proliferative capacity. Cancer Res. 2016, 76, 150–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durand, R.E.; Olive, P.L. Resistance of tumor cells to chemo- and radiotherapy modulated by the three-dimensional architecture of solid tumors and spheroids. Methods Cell Biol. 2001, 64, 211–233. [Google Scholar] [CrossRef]
- Friedrich, J.; Seidel, C.; Ebner, R.; Kunz-Schughart, L.A. Spheroid-based drug screen: Considerations and practical approach. Nat. Protoc. 2009, 4, 309–324. [Google Scholar] [CrossRef]
- Lazaro, C.A.; Croager, E.J.; Mitchell, C.; Campbell, J.S.; Yu, C.; Foraker, J.; Rhim, J.A.; Yeoh, G.C.; Fausto, N. Establishment, characterization, and long-term maintenance of cultures of human fetal hepatocytes. Hepatology 2003, 38, 1095–1106. [Google Scholar] [CrossRef]
- Kuperwasser, C.; Chavarria, T.; Wu, M.; Magrane, G.; Gray, J.W.; Carey, L.; Richardson, A.; Weinberg, R.A. Reconstruction of functionally normal and malignant human breast tissues in mice. Proc. Natl. Acad. Sci. USA 2004, 101, 4966–4971. [Google Scholar] [CrossRef] [Green Version]
- Nadkarni, R.R.; Abed, S.; Draper, J.S. Organoids as a model system for studying human lung development and disease. Biochem. Biophys. Res. Commun. 2016, 473, 675–682. [Google Scholar] [CrossRef]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, R.H.; Jones, J.K.; Gabriel, A.; Lukies, R.E. A new colon carcinoma cell line (LIM1863) that grows as organoids with spontaneous differentiation into crypt-like structures in vitro. Cancer Res. 1987, 47, 2683–2689. [Google Scholar] [PubMed]
- Lebret, S.C.; Newgreen, D.F.; Thompson, E.W.; Ackland, M.L. Induction of epithelial to mesenchymal transition in PMC42-LA human breast carcinoma cells by carcinoma-associated fibroblast secreted factors. Breast Cancer Res. 2007, 9, R19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.; Van Es, J.H.; Van den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [Green Version]
- Boj, S.F.; Hwang, C.I.; Baker, L.A.; Chio, I.I.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015, 160, 324–338. [Google Scholar] [CrossRef] [Green Version]
- Sachs, N.; de Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 2018, 172, 373–386. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.H.N.; Siu, H.C.; Law, S.; Ho, S.L.; Yue, S.S.K.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Ma, S.; Lam, K.O.; et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell 2018, 23, 882–897. [Google Scholar] [CrossRef] [Green Version]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Bottinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38. [Google Scholar] [CrossRef]
- Li, X.; Francies, H.E.; Secrier, M.; Perner, J.; Miremadi, A.; Galeano-Dalmau, N.; Barendt, W.J.; Letchford, L.; Leyden, G.M.; Goffin, E.K.; et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat. Commun. 2018, 9, 2983. [Google Scholar] [CrossRef]
- Mullenders, J.; de Jongh, E.; Brousali, A.; Roosen, M.; Blom, J.P.A.; Begthel, H.; Korving, J.; Jonges, T.; Kranenburg, O.; Meijer, R.; et al. Mouse and human urothelial cancer organoids: A tool for bladder cancer research. Proc. Natl. Acad. Sci. USA 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schutgens, F.; Rookmaaker, M.B.; Margaritis, T.; Rios, A.; Ammerlaan, C.; Jansen, J.; Gijzen, L.; Vormann, M.; Vonk, A.; Viveen, M.; et al. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat. Biotechnol. 2019, 37, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Knutsdottir, H.; Hui, K.; Weiss, M.J.; He, J.; Philosophe, B.; Cameron, A.M.; Wolfgang, C.L.; Pawlik, T.M.; Ghiaur, G.; et al. Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI Insight 2019, 4, e121490. [Google Scholar] [CrossRef] [PubMed]
- Rolver, M.G.; Elingaard-Larsen, L.O.; Pedersen, S.F. Assessing cell viability and death in 3D spheroid cultures of cancer cells. J. Vis. Exp. 2019. [Google Scholar] [CrossRef] [Green Version]
- Froehlich, K.; Haeger, J.D.; Heger, J.; Pastuschek, J.; Photini, S.M.; Yan, Y.; Lupp, A.; Pfarrer, C.; Mrowka, R.; Schleussner, E.; et al. Generation of multicellular breast cancer tumor spheroids: Comparison of different protocols. J. Mammary Gland Biol. Neoplasia 2016, 21, 89–98. [Google Scholar] [CrossRef]
- Foty, R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J. Vis. Exp. 2011. [Google Scholar] [CrossRef]
- Huang, B.W.; Gao, J.Q. Application of 3D cultured multicellular spheroid tumor models in tumor-targeted drug delivery system research. J. Control. Release 2018, 270, 246–259. [Google Scholar] [CrossRef]
- Cui, X.; Hartanto, Y.; Zhang, H. Advances in multicellular spheroids formation. J. R. Soc. Interface 2017, 14. [Google Scholar] [CrossRef]
- Nagelkerke, A.; Bussink, J.; Sweep, F.C.; Span, P.N. Generation of multicellular tumor spheroids of breast cancer cells: How to go three-dimensional. Anal. Biochem. 2013, 437, 17–19. [Google Scholar] [CrossRef]
- Costa, E.C.; Gaspar, V.M.; Coutinho, P.; Correia, I.J. Optimization of liquid overlay technique to formulate heterogenic 3D co-cultures models. Biotechnol. Bioeng. 2014, 111, 1672–1685. [Google Scholar] [CrossRef]
- Ivascu, A.; Kubbies, M. Rapid generation of single-tumor spheroids for high-throughput cell function and toxicity analysis. J. Biomol. Screen. 2006, 11, 922–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metzger, W.; Sossong, D.; Bachle, A.; Putz, N.; Wennemuth, G.; Pohlemann, T.; Oberringer, M. The liquid overlay technique is the key to formation of co-culture spheroids consisting of primary osteoblasts, fibroblasts and endothelial cells. Cytotherapy 2011, 13, 1000–1012. [Google Scholar] [CrossRef]
- Scherer, K.M.; Bisby, R.H.; Botchway, S.W.; Hadfield, J.A.; Haycock, J.W.; Parker, A.W. Three-dimensional imaging and uptake of the anticancer drug combretastatin in cell spheroids and photoisomerization in gels with multiphoton excitation. J. Biomed. Opt. 2015, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.Y.; Zou, J.Y.; Badar, M.; Liu, J.C.; Shi, W.T.; Wang, S.Q.; Guo, Q.Y.; Wang, X.F.; Kessel, S.; Chan, L.L.Y.; et al. Longitudinal morphological and physiological monitoring of three-dimensional tumor spheroids using optical coherence tomography. J. Vis. Exp. 2019. [Google Scholar] [CrossRef] [PubMed]
- Amann, A.; Zwierzina, M.; Gamerith, G.; Bitsche, M.; Huber, J.M.; Vogel, G.F.; Blumer, M.; Koeck, S.; Pechriggl, E.J.; Kelm, J.M.; et al. Development of an innovative 3D cell culture system to study tumour—Stroma interactions in non-small cell lung cancer cells. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, E.C.; de Melo-Diogo, D.; Moreira, A.F.; Carvalho, M.P.; Correia, I.J. Spheroids formation on non-adhesive surfaces by liquid overlay technique: Considerations and practical approaches. Biotechnol. J. 2018, 13. [Google Scholar] [CrossRef]
- Wartenberg, M.; Donmez, F.; Ling, F.C.; Acker, H.; Hescheler, J.; Sauer, H. Tumor-induced angiogenesis studied in confrontation cultures of multicellular tumor spheroids and embryoid bodies grown from pluripotent embryonic stem cells. FASEB J. 2001, 15, 995–1005. [Google Scholar] [CrossRef]
- Mazzoleni, G.; Di Lorenzo, D.; Steimberg, N. Modelling tissues in 3D: The next future of pharmaco-toxicology and food research? Genes Nutr. 2009, 4, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Del Duca, D.; Werbowetski, T.; Del Maestro, R.F. Spheroid preparation from hanging drops: Characterization of a model of brain tumor invasion. J. Neurooncol. 2004, 67, 295–303. [Google Scholar] [CrossRef]
- Li, Q.; Chen, C.; Kapadia, A.; Zhou, Q.; Harper, M.K.; Schaack, J.; LaBarbera, D.V. 3D models of epithelial-mesenchymal transition in breast cancer metastasis: High-throughput screening assay development, validation, and pilot screen. J. Biomol. Screen. 2011, 16, 141–154. [Google Scholar] [CrossRef] [Green Version]
- Hardelauf, H.; Frimat, J.P.; Stewart, J.D.; Schormann, W.; Chiang, Y.Y.; Lampen, P.; Franzke, J.; Hengstler, J.G.; Cadenas, C.; Kunz-Schughart, L.A.; et al. Microarrays for the scalable production of metabolically relevant tumour spheroids: A tool for modulating chemosensitivity traits. Lab Chip 2011, 11, 419–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Lyu, X.; Yi, M.; Zhao, W.; Song, Y.; Wu, K. Organoid technology and applications in cancer research. J. Hematol. Oncol. 2018, 11, 116. [Google Scholar] [CrossRef]
- Lin, M.; Gao, M.; Cavnar, M.J.; Kim, J. Utilizing gastric cancer organoids to assess tumor biology and personalize medicine. World J. Gastrointest. Oncol. 2019, 11, 509–517. [Google Scholar] [CrossRef]
- Nunes, A.S.; Barros, A.S.; Costa, E.C.; Moreira, A.F.; Correia, I.J. 3D tumor spheroids as in vitro models to mimic in vivo human solid tumors resistance to therapeutic drugs. Biotechnol. Bioeng. 2019, 116, 206–226. [Google Scholar] [CrossRef] [Green Version]
- Fiorini, E.; Veghini, L.; Corbo, V. Modeling cell communication in cancer with organoids: Making the complex simple. Front. Cell Dev. Biol. 2020, 8, 166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Sun, Q.; Pei, J. Microfluidic-based 3D engineered microvascular networks and their applications in vascularized microtumor models. Micromachines 2018, 9, 493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarvelainen, H.; Sainio, A.; Koulu, M.; Wight, T.N.; Penttinen, R. Extracellular matrix molecules: Potential targets in pharmacotherapy. Pharmacol. Rev. 2009, 61, 198–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, C.S.; Postovit, L.M.; Lajoie, G.A. Matrigel: A complex protein mixture required for optimal growth of cell culture. Proteomics 2010, 10, 1886–1890. [Google Scholar] [CrossRef]
- Broguiere, N.; Isenmann, L.; Hirt, C.; Ringel, T.; Placzek, S.; Cavalli, E.; Ringnalda, F.; Villiger, L.; Zullig, R.; Lehmann, R.; et al. Growth of epithelial organoids in a defined hydrogel. Adv. Mater. 2018, 30, e1801621. [Google Scholar] [CrossRef]
- Saheli, M.; Sepantafar, M.; Pournasr, B.; Farzaneh, Z.; Vosough, M.; Piryaei, A.; Baharvand, H. Three-dimensional liver-derived extracellular matrix hydrogel promotes liver organoids function. J. Cell Biochem. 2018, 119, 4320–4333. [Google Scholar] [CrossRef]
- Cruz-Acuna, R.; Quiros, M.; Farkas, A.E.; Dedhia, P.H.; Huang, S.; Siuda, D.; Garcia-Hernandez, V.; Miller, A.J.; Spence, J.R.; Nusrat, A.; et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 2017, 19, 1326–1335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gjorevski, N.; Sachs, N.; Manfrin, A.; Giger, S.; Bragina, M.E.; Ordonez-Moran, P.; Clevers, H.; Lutolf, M.P. Designer matrices for intestinal stem cell and organoid culture. Nature 2016, 539, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Fong, E.L.S.; Toh, T.B.; Lin, Q.X.X.; Liu, Z.; Hooi, L.; Rashid, M.; Benoukraf, T.; Chow, E.K.; Huynh, T.H.; Yu, H. Datasets describing the growth and molecular features of hepatocellular carcinoma patient-derived xenograft cells grown in a three-dimensional macroporous hydrogel. Data Brief 2018, 18, 594–606. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Shimokawa, M.; Date, S.; Takano, A.; Matano, M.; Nanki, K.; Ohta, Y.; Toshimitsu, K.; Nakazato, Y.; Kawasaki, K.; et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 2016, 18, 827–838. [Google Scholar] [CrossRef] [Green Version]
- Mazzucchelli, S.; Piccotti, F.; Allevi, R.; Truffi, M.; Sorrentino, L.; Russo, L.; Agozzino, M.; Signati, L.; Bonizzi, A.; Villani, L.; et al. Establishment and morphological characterization of patient-derived organoids from breast cancer. Biol. Proc. Online 2019, 21, 12. [Google Scholar] [CrossRef]
- Puca, L.; Bareja, R.; Prandi, D.; Shaw, R.; Benelli, M.; Karthaus, W.R.; Hess, J.; Sigouros, M.; Donoghue, A.; Kossai, M.; et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat. Commun. 2018, 9, 2404. [Google Scholar] [CrossRef] [Green Version]
- Fan, H.; Demirci, U.; Chen, P. Emerging organoid models: Leaping forward in cancer research. J. Hematol. Oncol. 2019, 12, 142. [Google Scholar] [CrossRef] [Green Version]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef]
- Kaushik, G.; Ponnusamy, M.P.; Batra, S.K. Concise review: Current status of three-dimensional organoids as preclinical models. Stem Cells 2018, 36, 1329–1340. [Google Scholar] [CrossRef] [Green Version]
- Nie, J.; Hashino, E. Organoid technologies meet genome engineering. EMBO Rep. 2017, 18, 367–376. [Google Scholar] [CrossRef]
- Broutier, L.; Mastrogiovanni, G.; Verstegen, M.M.; Francies, H.E.; Gavarro, L.M.; Bradshaw, C.R.; Allen, G.E.; Arnes-Benito, R.; Sidorova, O.; Gaspersz, M.P.; et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017, 23, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- Weeber, F.; van de Wetering, M.; Hoogstraat, M.; Dijkstra, K.K.; Krijgsman, O.; Kuilman, T.; Gadellaa-van Hooijdonk, C.G.; van der Velden, D.L.; Peeper, D.S.; Cuppen, E.P.; et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl. Acad. Sci. USA 2015, 112, 13308–13311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maru, Y.; Tanaka, N.; Ebisawa, K.; Odaka, A.; Sugiyama, T.; Itami, M.; Hippo, Y. Establishment and characterization of patient-derived organoids from a young patient with cervical clear cell carcinoma. Cancer Sci. 2019, 110, 2992–3005. [Google Scholar] [CrossRef] [PubMed]
- Nanki, K.; Toshimitsu, K.; Takano, A.; Fujii, M.; Shimokawa, M.; Ohta, Y.; Matano, M.; Seino, T.; Nishikori, S.; Ishikawa, K.; et al. Divergent routes toward Wnt and R-spondin niche independency during human gastric carcinogenesis. Cell 2018, 174, 856–869. [Google Scholar] [CrossRef] [Green Version]
- Maenhoudt, N.; Defraye, C.; Boretto, M.; Jan, Z.; Heremans, R.; Boeckx, B.; Hermans, F.; Arijs, I.; Cox, B.; Van Nieuwenhuysen, E.; et al. Developing organoids from ovarian cancer as experimental and preclinical models. Stem Cell Rep. 2020, 14, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Maru, Y.; Tanaka, N.; Itami, M.; Hippo, Y. Efficient use of patient-derived organoids as a preclinical model for gynecologic tumors. Gynecol. Oncol. 2019, 154, 189–198. [Google Scholar] [CrossRef]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef] [Green Version]
- Kenny, P.A.; Lee, G.Y.; Myers, C.A.; Neve, R.M.; Semeiks, J.R.; Spellman, P.T.; Lorenz, K.; Lee, E.H.; Barcellos-Hoff, M.H.; Petersen, O.W.; et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007, 1, 84–96. [Google Scholar] [CrossRef]
- Harma, V.; Virtanen, J.; Makela, R.; Happonen, A.; Mpindi, J.P.; Knuuttila, M.; Kohonen, P.; Lotjonen, J.; Kallioniemi, O.; Nees, M. A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS ONE 2010, 5, e10431. [Google Scholar] [CrossRef]
- Li, X.; Ootani, A.; Kuo, C. An air-liquid interface culture system for 3D organoid culture of diverse primary gastrointestinal tissues. Methods Mol. Biol. 2016, 1422, 33–40. [Google Scholar] [CrossRef]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid modeling of the tumor immune microenvironment. Cell 2018, 175, 1972–1988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papapetrou, E.P. Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nat. Med. 2016, 22, 1392–1401. [Google Scholar] [CrossRef]
- Navarro, A.M.; Susanto, E.; Falk, A.; Wilhelm, M. Modeling cancer using patient-derived induced pluripotent stem cells to understand development of childhood malignancies. Cell Death Discov. 2018, 4, 7. [Google Scholar] [CrossRef]
- Lee, D.F.; Su, J.; Kim, H.S.; Chang, B.; Papatsenko, D.; Zhao, R.Y.; Yuan, Y.; Gingold, J.; Xia, W.Y.; Darr, H.; et al. Modeling familial cancer with induced pluripotent stem cells. Cell 2015, 161, 240–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, C.T.; Wang, J.Y.; Lin, Y.F.; Wo, A.M.; Chen, B.P.C.; Lee, H. Three-dimensional spheroid culture targeting versatile tissue bioassays using a PDMS-based hanging drop array. Sci. Rep. 2017, 7, 4363. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.W.; Desterke, C.; Feraud, O.; Richard, S.; Ferlicot, S.; Verkarre, V.; Patard, J.J.; Loisel-Duwattez, J.; Foudi, A.; Griscelli, F.; et al. iPSC-derived cancer organoids recapitulate genomic and phenotypic alterations of c-Met-mutated hereditary kidney cancer. Cancer Res. 2019, 79 (Suppl. 13), 3688. [Google Scholar] [CrossRef]
- Ong, S.M.; Zhao, Z.; Arooz, T.; Zhao, D.; Zhang, S.; Du, T.; Wasser, M.; van Noort, D.; Yu, H. Engineering a scaffold-free 3D tumor model for in vitro drug penetration studies. Biomaterials 2010, 31, 1180–1190. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Man, S.; Graham, C.H.; Kapitain, S.J.; Teicher, B.A.; Kerbel, R.S. Acquired multicellular-mediated resistance to alkylating agents in cancer. Proc. Natl. Acad. Sci. USA 1993, 90, 3294–3298. [Google Scholar] [CrossRef] [Green Version]
- Lemmo, S.; Atefi, E.; Luker, G.D.; Tavana, H. Optimization of aqueous biphasic tumor spheroid microtechnology for anti-cancer drug testing in 3D culture. Cell Mol. Bioeng. 2014, 7, 344–354. [Google Scholar] [CrossRef]
- LaBonia, G.J.; Lockwood, S.Y.; Heller, A.A.; Spence, D.M.; Hummon, A.B. Drug penetration and metabolism in 3D cell cultures treated in a 3D printed fluidic device: Assessment of irinotecan via MALDI imaging mass spectrometry. Proteomics 2016, 16, 1814–1821. [Google Scholar] [CrossRef]
- Doublier, S.; Belisario, D.C.; Polimeni, M.; Annaratone, L.; Riganti, C.; Allia, E.; Ghigo, D.; Bosia, A.; Sapino, A. HIF-1 activation induces doxorubicin resistance in MCF7 3-D spheroids via P-glycoprotein expression: A potential model of the chemo-resistance of invasive micropapillary carcinoma of the breast. BMC Cancer 2012, 12, 4. [Google Scholar] [CrossRef] [Green Version]
- Pattni, B.S.; Nagelli, S.G.; Aryasomayajula, B.; Deshpande, P.P.; Kulkarni, A.; Hartner, W.C.; Thakur, G.; Degterev, A.; Torchilin, V.P. Targeting of micelles and liposomes loaded with the pro-apoptotic drug, NCL-240, into NCI/ADR-RES cells in a 3D spheroid model. Pharm. Res. 2016, 33, 2540–2551. [Google Scholar] [CrossRef]
- Halfter, K.; Hoffmann, O.; Ditsch, N.; Ahne, M.; Arnold, F.; Paepke, S.; Grab, D.; Bauerfeind, I.; Mayer, B. Testing chemotherapy efficacy in HER2 negative breast cancer using patient-derived spheroids. J. Transl. Med. 2016, 14, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeppesen, M.; Hagel, G.; Glenthoj, A.; Vainer, B.; Ibsen, P.; Harling, H.; Thastrup, O.; Jorgensen, L.N.; Thastrup, J. Short-term spheroid culture of primary colorectal cancer cells as an in vitro model for personalizing cancer medicine. PLoS ONE 2017, 12, e0183074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seidlitz, T.; Merker, S.R.; Rothe, A.; Zakrzewski, F.; von Neubeck, C.; Grutzmann, K.; Sommer, U.; Schweitzer, C.; Scholch, S.; Uhlemann, H.; et al. Human gastric cancer modelling using organoids. Gut 2019, 68, 207–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usui, T.; Sakurai, M.; Enjoji, S.; Kawasaki, H.; Umata, K.; Ohama, T.; Fujiwara, N.; Yabe, R.; Tsuji, S.; Yamawaki, H.; et al. Establishment of a novel model for anticancer drug resistance in three-dimensional primary culture of tumor microenvironment. Stem Cells Int. 2016, 2016, 7053872. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Flinders, C.; Mumenthaler, S.M.; Hummon, A.B. MALDI Mass spectrometry imaging for evaluation of therapeutics in colorectal tumor organoids. J. Am. Soc. Mass Spectrom. 2018, 29, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Frappart, P.O.; Walter, K.; Gout, J.; Beutel, A.K.; Morawe, M.; Arnold, F.; Breunig, M.; Barth, T.F.; Marienfeld, R.; Schulte, L.; et al. Pancreatic cancer-derived organoids—A disease modeling tool to predict drug response. Un. Eur. Gastroenterol. J. 2020, 8, 594–606. [Google Scholar] [CrossRef] [Green Version]
- Sharick, J.T.; Walsh, C.M.; Sprackling, C.M.; Pasch, C.A.; Pham, D.L.; Esbona, K.; Choudhary, A.; Garcia-Valera, R.; Burkard, M.E.; McGregor, S.M.; et al. Metabolic heterogeneity in patient tumor-derived organoids by primary site and drug treatment. Front. Oncol. 2020, 10, 553. [Google Scholar] [CrossRef]
- Verissimo, C.S.; Overmeer, R.M.; Ponsioen, B.; Drost, J.; Mertens, S.; Verlaan-Klink, I.; Gerwen, B.V.; van der Ven, M.; Wetering, M.V.; Egan, D.A.; et al. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. Elife 2016, 5, e18489. [Google Scholar] [CrossRef] [Green Version]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Nanki, Y.; Chiyoda, T.; Hirasawa, A.; Ookubo, A.; Itoh, M.; Ueno, M.; Akahane, T.; Kameyama, K.; Yamagami, W.; Kataoka, F.; et al. Patient-derived ovarian cancer organoids capture the genomic profiles of primary tumours applicable for drug sensitivity and resistance testing. Sci. Rep. 2020, 10, 12581. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Takeishi, K.; Li, Z.; Cervantes-Alvarez, E.; Collin de l’Hortet, A.; Guzman-Lepe, J.; Cui, X.; Zhu, J. Microenvironment of a tumor-organoid system enhances hepatocellular carcinoma malignancy-related hallmarks. Organogenesis 2017, 13, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Ehsan, S.M.; Welch-Reardon, K.M.; Waterman, M.L.; Hughes, C.C.; George, S.C. A three-dimensional in vitro model of tumor cell intravasation. Integr. Biol. 2014, 6, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Worsdorfer, P.; Dalda, N.; Kern, A.; Kruger, S.; Wagner, N.; Kwok, C.K.; Henke, E.; Ergun, S. Generation of complex human organoid models including vascular networks by incorporation of mesodermal progenitor cells. Sci. Rep. 2019, 9, 15663. [Google Scholar] [CrossRef] [Green Version]
- Chulpanova, D.S.; Kitaeva, K.V.; James, V.; Rizvanov, A.A.; Solovyeva, V.V. Therapeutic prospects of extracellular vesicles in cancer treatment. Front. Immunol. 2018, 9, 1534. [Google Scholar] [CrossRef] [Green Version]
- Shaimardanova, A.A.; Solovyeva, V.V.; Chulpanova, D.S.; James, V.; Kitaeva, K.V.; Rizvanov, A.A. Extracellular vesicles in the diagnosis and treatment of central nervous system diseases. Neural Regen. Res. 2020, 15, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Szvicsek, Z.; Oszvald, A.; Szabo, L.; Sandor, G.O.; Kelemen, A.; Soos, A.A.; Paloczi, K.; Harsanyi, L.; Tolgyes, T.; Dede, K.; et al. Extracellular vesicle release from intestinal organoids is modulated by Apc mutation and other colorectal cancer progression factors. Cell Mol. Life Sci. 2019, 76, 2463–2476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linkous, A.; Balamatsias, D.; Snuderl, M.; Edwards, L.; Miyaguchi, K.; Milner, T.; Reich, B.; Cohen-Gould, L.; Storaska, A.; Nakayama, Y.; et al. Modeling patient-derived glioblastoma with cerebral organoids. Cell Rep. 2019, 26, 3203–3211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pine, A.R.; Cirigliano, S.M.; Nicholson, J.G.; Hu, Y.; Linkous, A.; Miyaguchi, K.; Edwards, L.; Singhania, R.; Schwartz, T.H.; Ramakrishna, R.; et al. Tumor microenvironment is critical for the maintenance of cellular states found in primary glioblastomas. Cancer Discov. 2020, 10, 964–979. [Google Scholar] [CrossRef] [Green Version]
- Tang, M.; Xie, Q.; Gimple, R.C.; Zhong, Z.; Tam, T.; Tian, J.; Kidwell, R.L.; Wu, Q.L.; Prager, B.C.; Qiu, Z.X.; et al. Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res. 2020. [Google Scholar] [CrossRef] [PubMed]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernandez-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiriac, H.; Belleau, P.; Engle, D.D.; Plenker, D.; Deschenes, A.; Somerville, T.D.D.; Froeling, F.E.M.; Burkhart, R.A.; Denroche, R.E.; Jang, G.H.; et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 2018, 8, 1112–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruun, J.; Kryeziu, K.; Eide, P.W.; Moosavi, S.H.; Eilertsen, I.A.; Langerud, J.; Rosok, B.; Totland, M.Z.; Brunsell, T.H.; Pellinen, T.; et al. Patient-derived organoids from multiple colorectal cancer liver metastases reveal moderate intra-patient pharmacotranscriptomic heterogeneity. Clin. Cancer Res. 2020, 26, 4107–4119. [Google Scholar] [CrossRef] [Green Version]
- Sanmamed, M.F.; Chester, C.; Melero, I.; Kohrt, H. Defining the optimal murine models to investigate immune checkpoint blockers and their combination with other immunotherapies. Ann. Oncol. 2016, 27, 1190–1198. [Google Scholar] [CrossRef]
- Baker, K. Organoids provide an important window on inflammation in cancer. Cancers 2018, 10, 151. [Google Scholar] [CrossRef] [Green Version]
- Kopper, O.; de Witte, C.J.; Lohmussaar, K.; Valle-Inclan, J.E.; Hami, N.; Kester, L.; Balgobind, A.V.; Korving, J.; Proost, N.; Begthel, H.; et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Int. J. Gynecol. Cancer 2019, 29, A132–A133. [Google Scholar] [CrossRef]
- Lee, S.H.; Hu, W.H.; Matulay, J.T.; Silva, M.V.; Owczarek, T.B.; Kim, K.; Chua, C.W.; Barlow, L.J.; Kandoth, C.; Williams, A.B.; et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell 2018, 173, 515–528. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilazieva, Z.; Ponomarev, A.; Rutland, C.; Rizvanov, A.; Solovyeva, V. Promising Applications of Tumor Spheroids and Organoids for Personalized Medicine. Cancers 2020, 12, 2727. https://doi.org/10.3390/cancers12102727
Gilazieva Z, Ponomarev A, Rutland C, Rizvanov A, Solovyeva V. Promising Applications of Tumor Spheroids and Organoids for Personalized Medicine. Cancers. 2020; 12(10):2727. https://doi.org/10.3390/cancers12102727
Chicago/Turabian StyleGilazieva, Zarema, Aleksei Ponomarev, Catrin Rutland, Albert Rizvanov, and Valeriya Solovyeva. 2020. "Promising Applications of Tumor Spheroids and Organoids for Personalized Medicine" Cancers 12, no. 10: 2727. https://doi.org/10.3390/cancers12102727
APA StyleGilazieva, Z., Ponomarev, A., Rutland, C., Rizvanov, A., & Solovyeva, V. (2020). Promising Applications of Tumor Spheroids and Organoids for Personalized Medicine. Cancers, 12(10), 2727. https://doi.org/10.3390/cancers12102727