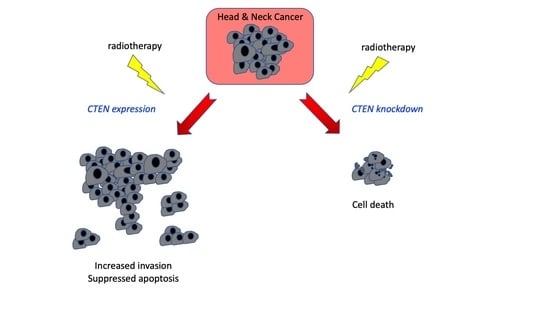
CTEN Induces Tumour Cell Invasion and Survival and Is Prognostic in Radiotherapy-Treated Head and Neck Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. CTEN Expression in HNSCC
2.2. CTEN Depletion Reduces Tumour Growth In Vivo
2.3. CTEN Supports Head and Neck Cancer Progression through Effects on Both Invasion and Survival
2.4. Loss of CTEN Expression Promotes Apoptosis
2.5. CTEN Is a Prognostic and Radiosensitivity Marker in HNSCC
3. Discussion
4. Materials and Methods
4.1. Antibodies and Reagents
4.2. Cell lines and Culture
4.3. Gene Silencing
4.4. Polymerase Chain Reaction (PCR)
4.5. Western Blot Analysis
4.6. Database Analysis
4.7. Cell Invasion Assay
4.8. Scratch Wound Assay
4.9. Colony Forming Assay
4.10. 3D Cell Proliferation Assay
4.11. Organotypic Culture
4.12. In Vivo Orthotopic Head and Neck Model
4.13. Apoptosis Protein Array
4.14. RNA Sequencing and Data Analysis
4.15. Tissue Microarry Array (TMA) Production and Immunohistochemistry
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Loyo, M.; Li, R.J.; Bettegowda, C.; Pickering, C.R.; Frederick, M.J.; Myers, J.N.; Agrawal, N. Lessons learned from next-generation sequencing in head and neck cancer. Head Neck 2013, 35, 454–463. [Google Scholar] [CrossRef]
- Brockstein, B.; Haraf, D.J.; Rademaker, A.W.; Kies, M.S.; Stenson, K.M.; Rosen, F.; Mittal, B.B.; Pelzer, H.; Fung, B.B.; Witt, M.E.; et al. Patterns of failure, prognostic factors and survival in locoregionally advanced head and neck cancer treated with concomitant chemoradiotherapy: A 9-year, 337-patient, multi-institutional experience. Ann. Oncol. 2004, 15, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Duray, A.; Lacremans, D.; Demoulin, S.; Delvenne, P.; Saussez, S. Prognosis of HPV-positive head and neck cancers: Implication of smoking and immunosuppression. Adv. Cell. Mol. Otolaryngol. 2014, 2. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Sougnez, C.; Lichtenstein, L.; Cibulskis, K.; Lander, E.; Gabriel, S.B.; Getz, G.; Ally, A.; Balasundaram, M.; Birol, I.; et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [Green Version]
- Cardesa, A.; Nadal, A. Carcinoma of the head and neck in the HPV era. Acta Dermatovenerol. Alp. Panonica. Adriat. 2011, 20, 161–173. [Google Scholar]
- Van Gysen, K.; Stevens, M.; Guo, L.; Jayamanne, D.; Veivers, D.; Wignall, A.; Pang, L.; Guminski, A.; Lee, A.; Hruby, G.; et al. Validation of the 8th edition UICC/AJCC TNM staging system for HPV associated oropharyngeal cancer patients managed with contemporary chemo-radiotherapy. BMC Cancer 2019, 19, e674. [Google Scholar] [CrossRef]
- 7th ed 2010 AJCC Cancer Staging Manual Seventh Edition. Available online: http://cancerstaging.org/references-tools/deskreferences/Documents/AJCC%207th%20Ed%20Cancer%20Staging%20Manual.pdf (accessed on 15 September 2020).
- Syrjänen, S. The role of human papillomavirus infection in head and neck cancers. Ann. Oncol. 2010, 21, vii243–vii245. [Google Scholar] [CrossRef]
- Fakhry, C.; Westra, W.H.; Li, S.; Cmelak, A.; Ridge, J.A.; Pinto, H.; Forastiere, A.; Gillison, M.L. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J. Natl. Cancer Inst. 2008, 100, 261–269. [Google Scholar] [CrossRef] [Green Version]
- O’Rorke, M.A.; Ellison, M.V.; Murray, L.J.; Moran, M.; James, J.; Anderson, L.A. Human papillomavirus related head and neck cancer survival: A systematic review and meta-analysis. Oral Oncol. 2012, 48, 1191–1201. [Google Scholar] [CrossRef]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tân, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef] [Green Version]
- AJCC Cancer Staging System, 8th Edition. Available online: https://www.springer.com/gp/book/9783319406176 (accessed on 17 September 2020).
- Krupar, R.; Robold, K.; Gaag, D.; Spanier, G.; Kreutz, M.; Renner, K.; Hellerbrand, C.; Hofstaedter, F.; Bosserhoff, A.K. Immunologic and metabolic characteristics of HPV-negative and HPV-positive head and neck squamous cell carcinomas are strikingly different. Virchows Arch. 2014, 465, 299–312. [Google Scholar] [CrossRef]
- Westra, W.H.; Taube, J.M.; Poeta, M.L.; Begum, S.; Sidransky, D.; Koch, W.M. Inverse relationship between human papillomavirus-16 infection and disruptive p53 gene mutations in squamous cell carcinoma of the head and neck. Clin. Cancer Res. 2008, 14, e2. [Google Scholar] [CrossRef] [Green Version]
- Adelstein, D.J.; Ridge, J.A.; Gillison, M.L.; Chaturvedi, A.K.; D’Souza, G.; Gravitt, P.E.; Westra, W.; Psyrri, A.; Kast, W.M.; Koutsky, L.A.; et al. Head and neck squamous cell cancer and the human papillomavirus: Summary of a National Cancer Institute State of the Science Meeting, November 9–10, 2008, Washington, D.C. Head Neck 2009, 31, 1393–1422. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.J.; Thirdborough, S.M.; Mellows, T.; Riley, C.; Harris, S.; Suchak, K.; Webb, A.; Hampton, C.; Patel, N.N.; Randall, C.J.; et al. Tumour-infiltrating lymphocytes predict for outcome in HPV-positive oropharyngeal cancer. Br. J. Cancer 2014, 110, 489–500. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, P.; Baujat, B.; Holostenco, V.; Bourredjem, A.; Baey, C.; Bourhis, J.; Pignon, J.P. Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): A comprehensive analysis by tumour site. Radiother. Oncol. 2011, 100, 33–40. [Google Scholar] [CrossRef]
- Cracchiolo, J.R.; Baxi, S.S.; Morris, L.G.; Ganly, I.; Patel, S.G.; Cohen, M.A.; Roman, B.R. Increase in primary surgical treatment of T1 and T2 oropharyngeal squamous cell carcinoma and rates of adverse pathologic features: National Cancer Data Base. Cancer 2016, 122, 1523–1532. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.H. C-terminal tensin-like (CTEN): A promising biomarker and target for cancer. Int. J. Biochem. Cell Biol. 2014, 51, 150–154. [Google Scholar] [CrossRef] [Green Version]
- Martuszewska, D.; Ljungberg, B.; Johansson, M.; Landberg, G.; Oslakovic, C.; Dahlbäck, B.; Hafizi, S. Tensin3 Is a Negative Regulator of Cell Migration and All Four Tensin Family Members Are Downregulated in Human Kidney Cancer. PLoS ONE 2009, 4, e13. [Google Scholar] [CrossRef]
- Qian, X.; Li, G.; Vass, W.; Papageorge, A.; Walker, R.C.; Asnaghi, L.; J, S.P.; Tosato, G.; Hunter, K.; Douglas R, L. The tensin-3 protein, including its SH2 domain, is phosphorylated by Src and contributes to tumorigenesis and metastasis. Cancer Cell 2009, 16, 246–258. [Google Scholar] [CrossRef] [Green Version]
- Katz, M.; Amit, I.; Citri, A.; Shay, T.; Carvalho, S.; Lavi, S.; Milanezi, F.; Lyass, L.; Amariglio, N.; Jacob-Hirsch, J.; et al. A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat. Cell Biol. 2007, 9, 961–969. [Google Scholar] [CrossRef]
- Albasri, A.; Aleskandarany, M.; Benhasouna, A.; Powe, D.G.; Ellis, I.O.; Ilyas, M.; Green, A.R. CTEN (C-terminal tensin-like), a novel oncogene overexpressed in invasive breast carcinoma of poor prognosis. Breast Cancer Res. Treat. 2011, 126, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Sjoestroem, C.; Khosravi, S.; Zhang, G.; Martinka, M.; Li, G. C-terminal tensin-like protein is a novel prognostic marker for primary melanoma patients. PLoS ONE 2013, 8, e80492. [Google Scholar] [CrossRef]
- Li, Y.; Mizokami, A.; Izumi, K.; Narimoto, K.; Shima, T.; Zhang, J.; Dai, J.; Keller, E.T.; Namiki, M. CTEN/tensin 4 expression induces sensitivity to paclitaxel in prostate cancer. Prostate 2010, 70, 48–60. [Google Scholar] [CrossRef] [Green Version]
- Albasri, A.; Al-Ghamdi, S.; Fadhil, W.; Aleskandarany, M.; Liao, Y.-C.; Jackson, D.; Lobo, D.N.; Lo, S.H.; Kumari, R.; Durrant, L.; et al. Cten signals through integrin-linked kinase (ILK) and may promote metastasis in colorectal cancer. Oncogene 2011, 30, 2997–3002. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.-Y.; Shih, Y.-P.; Li, T.; Carraway, K.L.; Lo, S.H. CTEN prolongs signaling by EGFR through reducing its ligand-induced degradation. Cancer Res. 2013, 73, 5266–5276. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.-C.; Si, L.; deVere White, R.W.; Lo, S.H. The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J. Cell Biol. 2007, 176, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Chan, L.-K.; Ko, F.C.F.; Ng, I.O.-L.; Yam, J.W.P. Deleted in liver cancer 1 (DLC1) utilizes a novel binding site for Tensin2 PTB domain interaction and is required for tumor-suppressive function. PLoS ONE 2009, 4, e5572. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Faisal, M.; Bakar, M.A.; Sarwar, A.; Adeel, M.; Batool, F.; Malik, K.I.; Jamshed, A.; Hussain, R. Depth of invasion (DOI) as a predictor of cervical nodal metastasis and local recurrence in early stage squamous cell carcinoma of oral tongue (ESSCOT). PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Almangush, A.; Bello, I.O.; Keski-Säntti, H.; Mäkinen, L.K.; Kauppila, J.H.; Pukkila, M.; Hagström, J.; Laranne, J.; Tommola, S.; Nieminen, O.; et al. Depth of invasion, tumor budding, and worst pattern of invasion: Prognostic indicators in early-stage oral tongue cancer. Head Neck 2014, 36, 811–818. [Google Scholar] [CrossRef]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P.; et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007, 9, 166–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, C.H.; Liao, C.T.; Peng, S.C.; Chen, Y.J.; Cheng, A.J.; Juang, J.L.; Tsai, C.Y.; Chen, T.C.; Chuang, Y.J.; Tang, C.Y.; et al. A novel molecular signature identified by systems genetics approach predicts prognosis in oral squamous cell carcinoma. PLoS ONE 2011, 6, e23452. [Google Scholar] [CrossRef] [PubMed]
- Ginos, M.A.; Page, G.P.; Michalowicz, B.S.; Patel, K.J.; Volker, S.E.; Pambuccian, S.E.; Ondrey, F.G.; Adams, G.L.; Gaffney, P.M. Identification of a Gene Expression Signature Associated with Recurrent Disease in Squamous Cell Carcinoma of the Head and Neck. Cancer Res. 2004, 64, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Pyeon, D.; Newton, M.A.; Lambert, P.F.; Den Boon, J.A.; Sengupta, S.; Marsit, C.J.; Woodworth, C.D.; Connor, J.P.; Haugen, T.H.; Smith, E.M.; et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007, 67. [Google Scholar] [CrossRef] [Green Version]
- ImageJ. Available online: https://imagej.nih.gov/ij/ (accessed on 3 June 2019).
- Gebäck, T.; Schulz, M.M.P.; Koumoutsakos, P.; Detmar, M. TScratch: A novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques 2009, 46, 265–274. [Google Scholar] [CrossRef]
- Guzmán, C.; Bagga, M.; Kaur, A.; Westermarck, J.; Abankwa, D. ColonyArea: An ImageJ plugin to automatically quantify colony formation in clonogenic assays. PLoS ONE 2014, 9, e92444. [Google Scholar] [CrossRef]
- Marsh, D.; Dickinson, S.; Neill, G.W.; Marshall, J.F.; Hart, I.R.; Thomas, G.J. v6 Integrin Promotes the Invasion of Morphoeic Basal Cell Carcinoma through Stromal Modulation. Cancer Res. 2008, 68, 3295–3303. [Google Scholar] [CrossRef] [Green Version]
- Ow, T.J.; Thomas, C.; Fulcher, C.D.; Chen, J.; López, A.; Reyna, D.E.; Prystowsky, M.B.; Smith, R.V.; Schiff, B.A.; Rosenblatt, G.; et al. Apoptosis signaling molecules as treatment targets in head and neck squamous cell carcinoma. Laryngoscope 2020. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, el1. [Google Scholar] [CrossRef] [Green Version]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, H.; Moriyama, S.; Mizuno, K.; Yukiue, H.; Konishi, A.; Yano, M.; Kaji, M.; Fukai, I.; Kiriyama, M.; Yamakawa, Y.; et al. Cten mRNA expression was correlated with tumor progression in lung cancers. Lung Cancer 2003, 40, 271–274. [Google Scholar] [CrossRef]
- Sakashita, K.; Mimori, K.; Tanaka, F.; Kamohara, Y.; Inoue, H.; Sawada, T.; Hirakawa, K.; Mori, M. Prognostic relevance of Tensin4 expression in human gastric cancer. Ann. Surg. Oncol. 2008, 15, 2606–2613. [Google Scholar] [CrossRef]
- Al-Ghamdi, S.; Cachat, J.; Albasri, A.; Ahmed, M.; Jackson, D.; Zaitoun, A.; Guppy, N.; Otto, W.R.; Alison, M.R.; Kindle, K.B.; et al. C-terminal tensin-like gene functions as an oncogene and promotes cell motility in pancreatic cancer. Pancreas 2013, 42, 135–140. [Google Scholar] [CrossRef]
- Liao, Y.-C.; Chen, N.-T.; Shih, Y.-P.; Dong, Y.; Lo, S.H. Up-regulation of C-terminal tensin-like molecule promotes the tumorigenicity of colon cancer through β-catenin. Cancer Res. 2009, 69, 4563–4566. [Google Scholar] [CrossRef] [Green Version]
- Al-Ghamdi, S.; Albasri, A.; Cachat, J.; Ibrahem, S.; Muhammad, B.A.; Jackson, D.; Nateri, A.S.; Kindle, K.B.; Ilyas, M. Cten Is Targeted by Kras Signalling to Regulate Cell Motility in the Colon and Pancreas. PLoS ONE 2011, 6, e8. [Google Scholar] [CrossRef]
- Hung, S.-Y.; Shih, Y.-P.; Chen, M.; Lo, S.H. Up-regulated cten by FGF2 contributes to FGF2-mediated cell migration. Mol. Carcinog. 2014, 53, 787–792. [Google Scholar] [CrossRef] [Green Version]
- Chan, L.-K.; Chiu, Y.-T.; Sze, K.M.-F.; Ng, I.O.-L. Tensin4 is up-regulated by EGF-induced ERK1/2 activity and promotes cell proliferation and migration in hepatocellular carcinoma. Oncotarget 2015, 6, 20964–20976. [Google Scholar] [CrossRef] [Green Version]
- Muharram, G.; Sahgal, P.; Korpela, T.; De Franceschi, N.; Kaukonen, R.; Clark, K.; Tulasne, D.; Carpén, O.; Ivaska, J. Tensin-4-dependent MET stabilization is essential for survival and proliferation in carcinoma cells. Dev. Cell 2014, 29, 421–436. [Google Scholar] [CrossRef] [Green Version]
- Lo, S.-S.; Lo, S.H.; Lo, S.H. Cleavage of cten by caspase-3 during apoptosis. Oncogene 2005, 24, 4311–4314. [Google Scholar] [CrossRef] [Green Version]
- Thorpe, H.; Asiri, A.; Akhlaq, M.; Ilyas, M. Cten promotes epithelial-mesenchymal transition through the post-transcriptional stabilization of Snail. Mol. Carcinog. 2017, 56, 2601–2609. [Google Scholar] [CrossRef] [PubMed]
- Franco, D.L.; Mainez, J.; Vega, S.; Sancho, P.; Murillo, M.M.; de Frutos, C.A.; Del Castillo, G.; López-Blau, C.; Fabregat, I.; Nieto, M.A. Snail1 suppresses TGF-beta-induced apoptosis and is sufficient to trigger EMT in hepatocytes. J. Cell Sci. 2010, 123, 3467–3477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedl, S.J.; Shi, Y. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 2004, 5, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.S.Y. Apoptosis in cancer: From pathogenesis to treatment. J. Exp. Clin. Cancer Res. 2011, 30, e87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, D.; Stigbrand, T. Radiation-induced cell death mechanisms. Tumor Biol. 2010, 31, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Shi, Z.; Zhao, S.; Wang, F.T.; Zhou, T.T.; Liu, B.; Bao, J.K. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012, 45, 487–498. [Google Scholar] [CrossRef]
- Palumbo, S.; Comincini, S. Autophagy and ionizing radiation in tumors: The “survive or not survive” dilemma. J. Cell. Physiol. 2013, 228, 1–8. [Google Scholar] [CrossRef]
- Datta, R.; Kojima, H.; Banach, D.; Bump, N.J.; Talanian, R.V.; Alnemri, E.S.; Weichselbaum, R.R.; Wong, W.W.; Kufe, D.W. Activation of a CrmA-insensitive, p35-sensitive pathway in ionizing radiation-induced apoptosis. J. Biol. Chem. 1997, 272, 1965–1969. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Little, J.B. p53 is involved in but not required for ionizing radiation-induced caspase-3 activation and apoptosis in human lymphoblast cell lines. Cancer Res. 1998, 58, 4277–4281. [Google Scholar]
- Rahmanian, N.; Hosseinimehr, S.J.; Khalaj, A. The paradox role of caspase cascade in ionizing radiation therapy. J. Biomed. Sci. 2016, 23. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Liu, Z.; Myers, J.N. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. J. Cell. Biochem. 2016, 117, 2682–2692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porter, A.G.; Jänicke, R.U. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef]
- Michelin, S.; Perez, M.D.R.; Dubner, D.; Gisone, P. Increased activity and involvement of caspase-3 in radiation-induced apoptosis in neural cells precursors from developing rat brain. Neurotoxicology 2004, 25, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Flores, E.R.; Tsai, K.Y.; Crowley, D.; Sengupta, S.; Yang, A.; McKeon, F.; Jacks, T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 2002, 416, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Rheinwald, J.G.; Beckett, M.A. Tumorigenic Keratinocyte Lines Requiring Anchorage and Fibroblast Support Cultured from Human Squamous Cell Carcinomas. Cancer Res. 1981, 41, 1657–1663. [Google Scholar] [PubMed]
- Frank, C.J.; McClatchey, K.D.; Devaney, K.O.; Carey, T.E. Evidence that loss of chromosome 18q is associated with tumor progression. Cancer Res. 1997, 57, 824–827. [Google Scholar] [PubMed]
- Lin, C.J.; Grandis, J.R.; Carey, T.E.; Gollin, S.M.; Whiteside, T.L.; Koch, W.M.; Ferris, R.L.; Lai, S.Y. Head and neck squamous cell carcinoma cell lines: Established models and rationale for selection. Head Neck 2007, 29, 163–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, J.C.; Graham, M.P.; Kumar, B.; Lindsay, M.; Kupfer, R.; Lyons, R.H.; Bradford, C.R.; Carey, T.E. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck 2012, 32, 417–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, W.D.; Stulberg, C.S.; Simpson, W.F. A permanent heteroploid human cell line with type B glucose-6-phosphate dehydrogenase. Proc. Soc. Exp. Biol. Med. 1971, 136, 1187–1191. [Google Scholar] [CrossRef] [PubMed]
- Brooks, S.C.; Locke, E.R.; Soule, H.D. Estrogen receptor in a human cell line (MCF 7) from breast carcinoma. J. Biol. Chem. 1973, 248, 6251–6253. [Google Scholar]
- Prime, S.S.; Nixon, S.V.; Crane, I.J.; Stone, A.; Matthews, J.B.; Maitland, N.J.; Remnant, L.; Powell, S.K.; Game, S.M.; Scully, C. The behaviour of human oral squamous cell carcinoma in cell culture. J. Pathol. 1990, 160, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.A.; Trask, D.K.; Kumar, B.; Los, G.; Castro, J.; Lee, J.S.-J.; Chen, J.; Wang, S.; Bradford, C.R.; Carey, T.E. Reversal of cisplatin resistance with a BH3 mimetic, (-)-gossypol, in head and neck cancer cells: Role of wild-type p53 and Bcl-xL. Mol. Cancer Ther. 2005, 4, 1096–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, G.J.; Lewis, M.P.; Whawell, S.A.; Russell, A.; Sheppard, D.; Hart, I.R.; Speight, P.M.; Marshall, J.F. Expression of the alphavbeta6 integrin promotes migration and invasion in squamous carcinoma cells. J. Investig. Dermatol. 2001, 117, 67–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, Q.; Dawson, M.I.; Soprano, D.R.; Soprano, K.J. Modulation of retinoic acid receptor function alters the growth inhibitory response of oral SCC cells to retinoids. Oncogene 2000, 19, 1457–1465. [Google Scholar] [CrossRef] [Green Version]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Moutasim, K.A.; Nystrom, M.L.; Thomas, G.J. Cell migration and invasion assays. Methods Mol. Biol. 2011, 731, 333–343. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Myers, J.N.; Holsinger, F.C.; Jasser, S.A.; Bekele, B.N.; Fidler, I.J. An orthotopic nude mouse model of oral tongue squamous cell carcinoma. Clin. Cancer Res. 2002, 8, 293–298. [Google Scholar]
- Estimating Tumour Volume in a Primary Orthotopic Mouse Model of Human Pancreatic Cancer Using Rapid Acquisition Magnetic Resonance Imaging. Available online: http://www.vipoa.org/cancer (accessed on 14 September 2020).
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2014, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57. [Google Scholar] [CrossRef]





| Category | All OPSCC | HPV-Positive OPSCC | HPV-Negative OPSCC | |||
|---|---|---|---|---|---|---|
| Frequency | % | Frequency | % | Frequency | % | |
| Final HPV Status | ||||||
| Negative | 113 | 43.6 | - | - | - | - |
| Positive | 146 | 56.4 | - | - | - | - |
| Gender | ||||||
| Female | 67 | 25.9 | 36 | 24.7 | 31 | 27.4 |
| Male | 192 | 74.1 | 110 | 75.3 | 82 | 72.6 |
| Age at Diagnosis | ||||||
| <50 | 54 | 20.8 | 39 | 26.7 | 15 | 13.3 |
| 50–69 | 156 | 60.2 | 88 | 60.3 | 68 | 60.2 |
| 70+ | 49 | 18.9 | 19 | 13.0 | 30 | 26.5 |
| Smoking | ||||||
| Non-smoker/Ex-smoker | 102 | 48.6 | 78 | 62.9 | 24 | 27.9 |
| Current smoker | 108 | 51.4 | 46 | 37.1 | 62 | 72.1 |
| Alcohol | ||||||
| Non-drinker/Ex drinker | 30 | 15.5 | 18 | 16.1 | 12 | 14.8 |
| Current drinker | 163 | 84.5 | 94 | 83.9 | 69 | 85.2 |
| Overall Stage | ||||||
| I/II | 45 | 17.5 | 11 | 7.6 | 34 | 30.4 |
| III/IV | 212 | 82.5 | 134 | 92.4 | 78 | 69.6 |
| T Stage | ||||||
| Tis/T1/T2 | 156 | 64.2 | 99 | 69.2 | 57 | 57 |
| T3/T4 | 86 | 35.4 | 43 | 30.1 | 43 | 43 |
| Tx | 1 | 0.4 | 1 | 0.7 | 0 | 0 |
| Nodal Metastases | ||||||
| No | 51 | 21.1 | 12 | 8.5 | 39 | 39 |
| Yes | 191 | 78.9 | 130 | 91.5 | 61 | 61 |
| N Stage | ||||||
| N0–N2a | 81 | 33.5 | 33 | 23.2 | 48 | 48 |
| N2b–N3 | 161 | 66.5 | 109 | 76.8 | 52 | 52 |
| Distant Mets at Presentation | ||||||
| No | 239 | 98.8 | 141 | 99.3 | 98 | 98 |
| Yes | 3 | 1.2 | 1 | 0.7 | 2 | 2 |
| Tumour Grade | ||||||
| Well/moderately differentiated | 88 | 34 | 29 | 19.9 | 59 | 52.2 |
| Poorly differentiated | 171 | 66 | 117 | 80.1 | 54 | 47.8 |
| Primary Treatment | ||||||
| Surgery | 97 | 37.5 | 52 | 35.6 | 45 | 33.1 |
| Neoadjuvant chemotherapy, surgery | 5 | 1.9 | 4 | 2.7 | 1 | 1.7 |
| Radiotherapy | 52 | 20.1 | 22 | 15.1 | 30 | 17.8 |
| Chemoradiotherapy | 89 | 34.4 | 64 | 43.8 | 25 | 30.4 |
| None/Palliative | 16 | 6.2 | 4 | 2.7 | 12 | 5.5 |
| Surgical Treatment Breakdown | ||||||
| Surgery only | 20 | 20.8 | 4 | 7.7 | 16 | 35.6 |
| Surgery, PORT | 68 | 70.8 | 41 | 78.8 | 28 | 62.2 |
| Surgery, POCRT | 8 | 8.3 | 7 | 13.5 | 1 | 2.2 |
| Margin Status | ||||||
| Negative | 61 | 61 | 29 | 60.4 | 28 | 60.9 |
| Close | 21 | 21 | 11 | 22.9 | 9 | 19.6 |
| Positive | 18 | 18 | 8 | 16.7 | 9 | 19.6 |
| CTEN Score | ||||||
| Absent/low | 115 | 44.4 | 86 | 58.9 | 29 | 25.7 |
| Moderate/high | 144 | 55.6 | 60 | 41.1 | 84 | 74.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fleming, J.C.; Woo, J.; Moutasim, K.; Hanley, C.J.; Frampton, S.J.; Wood, O.; Ward, M.; Woelk, C.H.; Ottensmeier, C.H.; Hafizi, S.; et al. CTEN Induces Tumour Cell Invasion and Survival and Is Prognostic in Radiotherapy-Treated Head and Neck Cancer. Cancers 2020, 12, 2963. https://doi.org/10.3390/cancers12102963
Fleming JC, Woo J, Moutasim K, Hanley CJ, Frampton SJ, Wood O, Ward M, Woelk CH, Ottensmeier CH, Hafizi S, et al. CTEN Induces Tumour Cell Invasion and Survival and Is Prognostic in Radiotherapy-Treated Head and Neck Cancer. Cancers. 2020; 12(10):2963. https://doi.org/10.3390/cancers12102963
Chicago/Turabian StyleFleming, Jason C., Jeongmin Woo, Karwan Moutasim, Christopher J. Hanley, Steven J. Frampton, Oliver Wood, Matthew Ward, Christopher H. Woelk, Christian H. Ottensmeier, Sassan Hafizi, and et al. 2020. "CTEN Induces Tumour Cell Invasion and Survival and Is Prognostic in Radiotherapy-Treated Head and Neck Cancer" Cancers 12, no. 10: 2963. https://doi.org/10.3390/cancers12102963
APA StyleFleming, J. C., Woo, J., Moutasim, K., Hanley, C. J., Frampton, S. J., Wood, O., Ward, M., Woelk, C. H., Ottensmeier, C. H., Hafizi, S., Kim, D., & Thomas, G. J. (2020). CTEN Induces Tumour Cell Invasion and Survival and Is Prognostic in Radiotherapy-Treated Head and Neck Cancer. Cancers, 12(10), 2963. https://doi.org/10.3390/cancers12102963