Enhancement of Breast Cancer Cell Aggressiveness by lncRNA H19 and its Mir-675 Derivative: Insight into Shared and Different Actions
Abstract
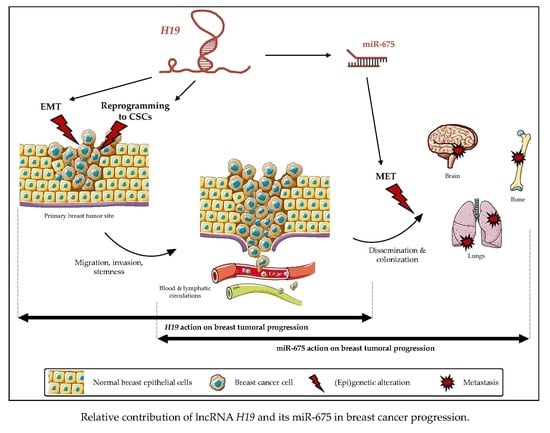
:1. Introduction
2. Results
2.1. LncRNA H19 and miR-675 Promote Breast Cancer Cell Invasion in Zebrafish Xenograft Model
2.2. LncRNA H19 and miR-675 Enhance Breast Cancer Cell Migration and Invasion in Collagen Gel
2.3. LncRNA H19 and miR-675 Differentially Regulate the Expression of Epithelial and Mesenchymal Markers
2.4. LncRNA H19 and miR-675 Promote Breast Cancer Colony Formation
2.5. LncRNA H19 and miR-675 Enhance the Stemness Properties of Breast Cancer Cells
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Establishment of H19 and miR-675 Overexpressing Cell Lines
4.3. siRNA and miRNA Transfection
4.4. RNA Extraction, Reverse Transcription and Real-time RT-PCR
4.5. Migration and Invasion Assays
4.6. Colony-forming and Sphere-forming Capacities
4.7. Aldefluor Assay
4.8. Western Blot Analysis
4.9. Transgenic Zebrafish Xenograft
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Angrand, P.O.; Vennin, C.; Le Bourhis, X.; Adriaenssens, E. The role of long non-coding RNAs in genome formatting and expression. Front Genet. 2015, 6, e165. [Google Scholar] [CrossRef] [Green Version]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adriaenssens, E.; Dumont, L.; Lottin, S.; Bolle, D.; Leprêtre, A.; Delobelle, A.; Bouali, F.; Dugimont, T.; Coll, J.; Curgy, J.J. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am. J. Pathol. 1998, 153, 1597–1607. [Google Scholar] [CrossRef] [Green Version]
- Adriaenssens, E.; Lottin, S.; Dugimont, T.; Fauquette, W.; Coll, J.; Dupouy, J.P.; Boilly, B.; Curgy, J.J. Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene 1999, 18, 4460–4473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Kahri, A.I.; Heikkilä, P.; Ilvesmäki, V.; Voutilainen, R. H19 and insulin-like growth factor-II gene expression in adrenal tumors and cultured adrenal cells. J. Clin. Endocrinol. Metab. 1995, 80, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Ariel, I.; Weinstein, D.; Voutilainen, R.; Schneider, T.; Lustig-Yariv, O.; de Groot, N.; Hochberg, A. Genomic imprinting and the endometrial cycle. The expression of the imprinted gene H19 in the human female reproductive organs. Diagn Mol Pathol Am. J. Surg. Pathol. Part B 1997, 6, 17–25. [Google Scholar] [CrossRef]
- Liu, G.; Xiang, T.; Wu, Q.F.; Wang, W.X. Long Noncoding RNA H19-Derived miR-675 Enhances Proliferation and Invasion via RUNX1 in Gastric Cancer Cells; Cognizant Communication Corporation: Putnam Valley, NY, USA, 2016; Volume 23, pp. 99–107. [Google Scholar] [CrossRef]
- Liang, W.C.; Fu, W.M.; Wong, C.W.; Wang, Y.; Wang, W.M.; Hu, G.X.; Zhang, L.; Xiao, L.J.; Wan, D.C.; Zhang, J.F.; et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 2015, 6, 22513–22525. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Chen, Z.; Fang, J.; Xu, A.; Zhang, W.; Wang, Z. H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumor Biol. 2016, 37, 263–270. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Cai, Y.; Zhao, X.; Jia, X.; Zhang, J.; Liu, J.; Zhen, H.; Wang, T.; Tang, X.; Liu, Y.; et al. Down-regulated long non-coding RNA H19 inhibits carcinogenesis of renal cell carcinoma. Neoplasma 2015, 62, 412–418. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.; Luan, W.; Wang, P.; Tao, T.; Zhang, J.; Qian, J.; Liu, N.; You, Y. Long Non-Coding RNA H19 Promotes Glioma Cell Invasion by Deriving miR-675. PLoS ONE 2014, 9, e86295. [Google Scholar] [CrossRef] [Green Version]
- Berteaux, N.; Lottin, S.; Monté, D.; Pinte, S.; Quatannens, B.; Coll, J.; Hondermarck, H.; Curgy, J.J.; Dugimont, T.; Adriaenssens, E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J. Biol. Chem. 2005, 280, 29625–29636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecerf, C.; Le Bourhis, X.; Adriaenssens, E. The long non-coding RNA H19: An active player with multiple facets to sustain the hallmarks of cancer. Cell Mol. Life Sci. 2019, 76, 4673–4687. [Google Scholar] [CrossRef] [PubMed]
- Lottin, S.; Adriaenssens, E.; Dupressoir, T.; Berteaux, N.; Montpellier, C.; Coll, J.; Dugimont, T.; Curgy, J.J. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 2002, 23, 1885–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, X.; Zang, R.; Zhang, E.; Liu, Y.; Shi, X.; Zhang, E.; Shao, L.; Li, A.; Yang, N.; Han, X.; et al. LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget 2016, 7, 81452–81462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, F.; Li, T.T.; Wang, K.L.; Xiao, G.Q.; Wang, J.H.; Zhao, H.D.; Kang, Z.J.; Fan, W.J.; Zhu, L.L.; Li, M.; et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017, 8, e2569. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Li, Y.; Li, Y.; Ren, K.; Li, X.; Han, X.; Wang, J. Long non-coding RNA H19 promotes the proliferation and invasion of breast cancer through upregulating DNMT1 expression by sponging miR-152. J. Biochem. Mol. Toxicol. 2017, 31, e21933. [Google Scholar] [CrossRef]
- Cai, X.; Cullen, B.R. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007, 13, 313–316. [Google Scholar] [CrossRef] [Green Version]
- Garzon, R.; Fabbri, M.; Cimmino, A.; Calin, G.A.; Croce, C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006, 12, 580–587. [Google Scholar] [CrossRef]
- Tsang, W.P.; Ng, E.K.O.; Ng, S.S.M.; Jin, H.; Yu, J.; Sung, J.J.Y.; Kwok, T.T. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010, 31, 350–358. [Google Scholar] [CrossRef]
- Hernandez, J.M.; Elahi, A.; Clark, C.W.; Wang, J.; Humphries, L.A.; Centeno, B.; Bloom, G.; Fuchs, B.C.; Yeatman, T.; Shibata, D. miR-675 Mediates Downregulation of Twist1 and Rb in AFP-Secreting Hepatocellular Carcinoma. Ann Surg. Oncol. 2013, 20, 625–635. [Google Scholar] [CrossRef]
- Zhuang, M.; Gao, W.; Xu, J.; Wang, P.; Shu, Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem. Biophys. Res. Commun. 2014, 448, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Vennin, C.; Spruyt, N.; Dahmani, F.; Julien, S.; Bertucci, F.; Finetti, P.; Chassat, T.; Bourette, R.P.; Le Bourhis, X.; Adriaenssens, E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 2015, 6, 29209–29223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, D.; Chen, L.; Snaar-Jagalska, E.; Chaudhry, B. Embryonic zebrafish xenograft assay of human cancer metastasis. F1000Research 2018, 7, e1682. [Google Scholar] [CrossRef]
- Daulat, A.M.; Bertucci, F.; Audebert, S.; Sergé, A.; Finetti, P.; Josselin, E.; Castellano, R.; Birnbaum, D.; Angers, S.; Borg, J.P. PRICKLE1 Contributes to Cancer Cell Dissemination through Its Interaction with mTORC2. Dev. Cell 2016, 37, 311–325. [Google Scholar] [CrossRef]
- Charafe-Jauffret, E.; Ginestier, C.; Iovino, F.; Wicinski, J.; Cervera, N.; Finetti, P.; Hur, M.J.; Diebel, M.E.; Monville, F.; Dutcher, J.; et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009, 69, 1302–1313. [Google Scholar] [CrossRef] [Green Version]
- Creighton, C.J.; Li, X.; Landis, M.; Dixon, J.M.; Neumeister, V.M.; Sjolund, A.; Rimm, D.L.; Wong, H.; Rodriguez, A.; Herschkowitz, J.I.; et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc. Natl. Acad. Sci. USA 2009, 106, 13820–13825. [Google Scholar] [CrossRef] [Green Version]
- Ishiguro, T.; Ohata, H.; Sato, A.; Yamawaki, K.; Enomoto, T.; Okamoto, K. Tumor-derived spheroids: Relevance to cancer stem cells and clinical applications. Cancer Sci. 2017, 108, 283–289. [Google Scholar] [CrossRef] [Green Version]
- Grolez, G.P.; Hammadi, M.; Barras, A.; Gordienko, D.; Slomianny, C.; Völkel, P.; Angrand, P.O.; Pinault, M.; Guimaraes, C.; Potier-Cartereau, M.; et al. Encapsulation of a TRPM8 Agonist, WS12, in Lipid Nanocapsules Potentiates PC3 Prostate Cancer Cell Migration Inhibition through Channel Activation. Sci. Rep. 2019, 9, e7926. [Google Scholar] [CrossRef]
- Raby, L.; Völkel, P.; Le Bourhis, X.; Angrand, P.O. La transplantation de cellules tumorales chez le poisson zèbre: De la recherche translationnelle à la médecine personnalisée. Bull Cancer 2020, 107, 30–40. [Google Scholar] [CrossRef]
- Shima, H.; Kida, K.; Adachi, S.; Yamada, A.; Sugae, S.; Narui, K.; Miyagi, Y.; Nishi, M.; Ryo, A.; Murata, S.; et al. Lnc RNA H19 is associated with poor prognosis in breast cancer patients and promotes cancer stemness. Breast Cancer Res. Treat. 2018, 170, 507–516. [Google Scholar] [CrossRef]
- Liao, S.; Yu, C.; Liu, H.; Zhang, C.; Li, Y.; Zhong, X. Long non-coding RNA H19 promotes the proliferation and invasion of lung cancer cells and regulates the expression of E-cadherin, N-cadherin, and vimentin. OncoTargets Ther. 2019, 12, 4099–4107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matouk, I.J.; Halle, D.; Raveh, E.; Gilon, M.; Sorin, V.; Hochberg, A. The role of the oncofetal H19 lncRNA in tumor metastasis: Orchestrating the EMT-MET decision. Oncotarget 2016, 7, 3748–3765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, P.; Qiu, K.; Jia, Y. Modeling of mesenchymal hybrid epithelial state and phenotypic transitions in EMT and MET processes of cancer cells. Sci. Rep. 2018, 8, e14323. [Google Scholar] [CrossRef] [PubMed]
- Mayor, R.; Etienne-Manneville, S. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 2016, 17, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Satoh, K.; Hamada, S.; Shimosegawa, T. Involvement of epithelial to mesenchymal transition in the development of pancreatic ductal adenocarcinoma. J. Gastroenterol. 2015, 50, 140–146. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Gu, L.N.; Shan, B.E.; Geng, C.Z.; Sang, M.X. Biomarkers for EMT and MET in breast cancer: An update. Oncol. Lett. 2016, 12, 4869–4876. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.J.; Deng, J.; Chen, C.; Hu, W.; Yuan, Y.C.; Xia, Z.K. LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. Int. J. Biochem. Cell Biol. 2019, 113, 27–36. [Google Scholar] [CrossRef]
- Liu, L.; Liu, L.; Lu, S. lncRNA H19 promotes viability and epithelial-mesenchymal transition of lung adenocarcinoma cells by targeting miR-29b-3p and modifying STAT3. Int. J. Oncol. 2019, 54, 929–941. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.Q.; Zeng, D.; Chen, C.F.; Sun, S.M.; Lu, X.F.; Peng, C.; Lin, H.Y. Long noncoding RNA H19 is a critical oncogenic driver and contributes to epithelial-mesenchymal transition in papillary thyroid carcinoma. Cancer Manag. Res. 2019, 11, 2059–2072. [Google Scholar] [CrossRef] [Green Version]
- Xiong, H.; Shen, J.; Chen, Z.; Yang, J.; Xie, B.; Jia, Y.; Jayasinghe, U.; Wang, J.; Zhao, W.; Xie, S.; et al. H19/let-7/Lin28 ceRNA network mediates autophagy inhibiting epithelial-mesenchymal transition in breast cancer. Int. J. Oncol. 2020, 56, 794–806. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, F.; Yuan, J.; Yuan, S.; Zhou, W.; Huo, X.; Xu, D.; Bi, H.S.; Wang, F.; Sun, S.H. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 2013, 34, 577–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, M.; Zhong, Z.; Huang, M.; Tian, Q.; Jiang, R.; Chen, J. lncRNA H19 regulates epithelial–mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim Biophys Acta BBA. Mol. Cell. Res. 2017, 1864, 1887–1899. [Google Scholar] [CrossRef]
- O’Brien, S.J.; Carter, J.V.; Burton, J.F.; Oxford, B.G.; Schmidt, M.N.; Hallion, J.C.; Galandiuk, S. The role of the miR-200 family in epithelial–mesenchymal transition in colorectal cancer: A systematic review. Int. J. Cancer 2018, 142, 2501–2511. [Google Scholar] [CrossRef] [PubMed]
- Title, A.C.; Hong, S.J.; Pires, N.D.; Hasenöhrl, L.; Godbersen, S.; Stokar-Regenscheit, N.; Bartel, D.P.; Stoffel, M. Genetic dissection of the miR-200-Zeb1 axis reveals its importance in tumor differentiation and invasion. Nat. Commun. 2018, 9, e4671. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Li, M.; Chen, H.; Zhao, Y.; Gao, S.; Cheng, C. H19 Functions as a ceRNA in Promoting Metastasis Through Decreasing miR-200s Activity in Osteosarcoma. DNA Cell Biol. 2016, 35, 235–240. [Google Scholar] [CrossRef]
- Zhao, Y.; Feng, C.; Li, Y.; Ma, Y.; Cai, R. LncRNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol. Cell Biochem. 2019, 460, 1–8. [Google Scholar] [CrossRef]
- Zhou, S.; He, Y.; Yang, S.; Hu, J.; Zhang, Q.; Chen, W.; Xu, H.; Zhang, H.; Zhong, S.; Zhao, J.; et al. The regulatory roles of lncRNAs in the process of breast cancer invasion and metastasis. Biosci. Rep. 2018, 38, 1–2. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Li, Y.; Xiao, G.D.; Zheng, X.Q.; Wang, J.C.; Xu, C.W.; Qin, S.; Ren, H.; Tang, S.H.; Sun, X. H19 regulation of oestrogen induction of symmetric division is achieved by antagonizing Let-7c in breast cancer stem-like cells. Cell Prolif. 2019, 52, e12534. [Google Scholar] [CrossRef] [Green Version]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [Green Version]
- Tomita, H.; Tanaka, K.; Tanaka, T.; Hara, A. Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 2016, 7, 11018–11032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Chu, J.; Shen, X.; Wang, J.; Orkin, S.H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell 2008, 132, 1049–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hadjimichael, C.; Chanoumidou, K.; Papadopoulou, N.; Arampatzi, P.; Papamatheakis, J.; Kretsovali, A. Common stemness regulators of embryonic and cancer stem cells. World J. Stem Cells 2015, 7, 1150–1184. [Google Scholar] [CrossRef] [PubMed]
- Barsyte-Lovejoy, D.; Lau, S.K.; Boutros, P.C.; Khosravi, F.; Jurisica, I.; Andrulis, I.L.; Tsao, M.S.; Penn, L.Z. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006, 66, 5330–5337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abi Habib, W.; Azzi, S.; Brioude, F.; Steunou, V.; Thibaud, N.; Das Neves, C.; Le Jule, M.; Chantot-Bastaraud, S.; Keren, B.; Lyonnet, S.; et al. Extensive investigation of the IGF2/H19 imprinting control region reveals novel OCT4/SOX2 binding site defects associated with specific methylation patterns in Beckwith-Wiedemann syndrome. Hum Mol. Genet. 2014, 23, 5763–5773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Qiao, Y.; Han, D.; Zhang, Y.; Ma, N. Enemy or partner: Relationship between intronic micrornas and their host genes. IUBMB Life 2012, 64, 835–840. [Google Scholar] [CrossRef]
- Shi, W.; Gerster, K.; Alajez, N.M.; Tsang, J.; Waldron, L.; Pintilie, M.; Hui, A.B.; Sykes, J.; P’ng, C.; Miller, N.; et al. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011, 71, 2926–2937. [Google Scholar] [CrossRef] [Green Version]
- Qiao, Y.; Ma, N.; Wang, X.; Hui, Y.; Li, F.; Xiang, Y.; Zhou, J.; Zou, C.; Jin, J.; Lv, G.; et al. MiR-483-5p controls angiogenesis in vitro and targets serum response factor. FEBS Lett. 2011, 585, 3095–3100. [Google Scholar] [CrossRef]
- Collette, J.; Le Bourhis, X.; Adriaenssens, E. Regulation of Human Breast Cancer by the Long Non-Coding RNA H19. Int. J. Mol. Sci. 2017, 18, e2319. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Redpath, R.E.; Zhang, C.; Ning, N. Long non-coding RNA H19 promotes the migration and invasion of colon cancer cells via MAPK signaling pathway. Oncol. Lett. 2018, 16, 3365–3372. [Google Scholar] [CrossRef]
- Zhao, J.; Ma, S.T. Downregulation of lncRNA H19 inhibits migration and invasion of human osteosarcoma through the NF-κB pathway. Mol. Med. Rep. 2018, 17, 7388–7394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berteaux, N.; Aptel, N.; Cathala, G.; Genton, C.; Coll, J.; Daccache, A.; Spruyt, N.; Hondermarck, H.; Dugimont, T.; Curgy, J.J.; et al. A novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol. Cell Biol. 2008, 28, 6731–6745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lawson, N.D.; Weinstein, B.M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002, 248, 307–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]










© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peperstraete, E.; Lecerf, C.; Collette, J.; Vennin, C.; Raby, L.; Völkel, P.; Angrand, P.-O.; Winter, M.; Bertucci, F.; Finetti, P.; et al. Enhancement of Breast Cancer Cell Aggressiveness by lncRNA H19 and its Mir-675 Derivative: Insight into Shared and Different Actions. Cancers 2020, 12, 1730. https://doi.org/10.3390/cancers12071730
Peperstraete E, Lecerf C, Collette J, Vennin C, Raby L, Völkel P, Angrand P-O, Winter M, Bertucci F, Finetti P, et al. Enhancement of Breast Cancer Cell Aggressiveness by lncRNA H19 and its Mir-675 Derivative: Insight into Shared and Different Actions. Cancers. 2020; 12(7):1730. https://doi.org/10.3390/cancers12071730
Chicago/Turabian StylePeperstraete, Evodie, Clément Lecerf, Jordan Collette, Constance Vennin, Ludivine Raby, Pamela Völkel, Pierre-Olivier Angrand, Marie Winter, François Bertucci, Pascal Finetti, and et al. 2020. "Enhancement of Breast Cancer Cell Aggressiveness by lncRNA H19 and its Mir-675 Derivative: Insight into Shared and Different Actions" Cancers 12, no. 7: 1730. https://doi.org/10.3390/cancers12071730
APA StylePeperstraete, E., Lecerf, C., Collette, J., Vennin, C., Raby, L., Völkel, P., Angrand, P. -O., Winter, M., Bertucci, F., Finetti, P., Lagadec, C., Meignan, S., Bourette, R. P., Bourhis, X. L., & Adriaenssens, E. (2020). Enhancement of Breast Cancer Cell Aggressiveness by lncRNA H19 and its Mir-675 Derivative: Insight into Shared and Different Actions. Cancers, 12(7), 1730. https://doi.org/10.3390/cancers12071730