Loss of Tid1/DNAJA3 Co-Chaperone Promotes Progression and Recurrence of Hepatocellular Carcinoma after Surgical Resection: A Novel Model to Stratify Risk of Recurrence
Abstract
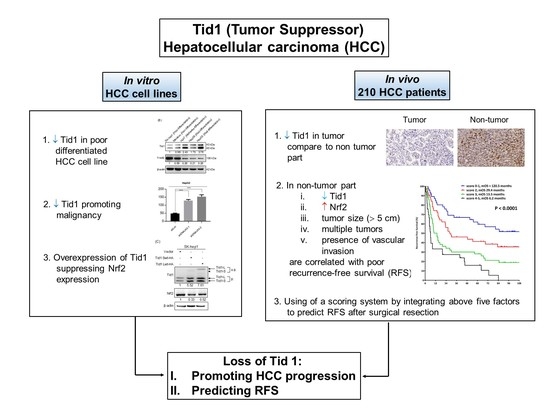
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Tid1 Functions as a Tumor Suppressor in HCC Cell Lines In Vitro
2.2. Expression Correlation between the Tid1 and Nrf2 Transcripts
2.3. Clinicopathological Features and Protein Level of Tid1 and Nrf2 of HBV-HCC and HCV-HCC
2.4. Tid1 Expression in Non-Tumor and Tumor Part of TNM-T Stage Specific HCC
2.5. Tid1 Expression in Non-Tumor Liver Tissues of Cirrhotic and Non-Cirrhotic State of HCC
2.6. Tid1 as a Prognostic Factor for Recurrence-Free Survival in HCC
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Human Specimens
4.3. Data Collection of DNAJA3 (Tid1) and NFEL2 (Nrf2) mRNA Expression from Open Source Database
4.4. Western Blotting
4.5. Overexpression by hTid1-L or -S Isoform
4.6. Downregulation by Small Hairpin RNA Interference (shRNAi)
4.7. Cell Anchorage-Independent Growth Assay
4.8. Immunohistochemistry (IHC)
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Ghouri, Y.A.; Mian, I.; Rowe, J.H. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J. Carcinog. 2017, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012, 142, 1264–1273.e1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poon, R.T.; Fan, S.T.; Lo, C.M.; Liu, C.L.; Wong, J. Long-term survival and pattern of recurrence after resection of small hepatocellular carcinoma in patients with preserved liver function: Implications for a strategy of salvage transplantation. Ann. Surg. 2002, 235, 373–382. [Google Scholar] [CrossRef]
- Lohitesh, K.; Chowdhury, R.; Mukherjee, S. Resistance a major hindrance to chemotherapy in hepatocellular carcinoma: An insight. Cancer Cell Int. 2018, 18, 44. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.C.; Hsu, C.; Chen, L.T.; Cheng, C.C.; Hu, F.C.; Cheng, A.L. Adjuvant interferon therapy after curative therapy for hepatocellular carcinoma (HCC): A meta-regression approach. J. Hepatol. 2010, 52, 889–894. [Google Scholar] [CrossRef]
- Singal, A.K.; Freeman, D.H., Jr.; Anand, B.S. Meta-analysis: Interferon improves outcomes following ablation or resection of hepatocellular carcinoma. Aliment. Pharmacol. Ther. 2010, 32, 851–858. [Google Scholar] [CrossRef]
- Yin, J.; Li, N.; Han, Y.; Xue, J.; Deng, Y.; Shi, J.; Guo, W.; Zhang, H.; Wang, H.; Cheng, S.; et al. Effect of antiviral treatment with nucleotide/nucleoside analogs on postoperative prognosis of hepatitis B virus-related hepatocellular carcinoma: A two-stage longitudinal clinical study. J. Clin. Oncol. 2013, 31, 3647–3655. [Google Scholar] [CrossRef]
- Kurzik-Dumke, U.; Gundacker, D.; Renthrop, M.; Gateff, E. Tumor suppression in Drosophila is causally related to the function of the lethal(2) tumorous imaginal discs gene, a dnaJ homolog. Dev. Genet. 1995, 16, 64–76. [Google Scholar] [CrossRef]
- Yin, X.; Rozakis-Adcock, M. Genomic organization and expression of the human tumorous imaginal disc (TID1) gene. Gene 2001, 278, 201–210. [Google Scholar] [CrossRef]
- Kurzik-Dumke, U.; Czaja, J. Htid-1, the human homolog of the Drosophila melanogaster l(2)tid tumor suppressor, defines a novel physiological role of APC. Cell Signal 2007, 19, 1973–1985. [Google Scholar] [CrossRef]
- Lo, J.F.; Hayashi, M.; Woo-Kim, S.; Tian, B.; Huang, J.F.; Fearns, C.; Takayama, S.; Zapata, J.M.; Yang, Y.; Lee, J.D. Tid1, a cochaperone of the heat shock 70 protein and the mammalian counterpart of the Drosophila tumor suppressor l(2)tid, is critical for early embryonic development and cell survival. Mol. Cell Biol. 2004, 24, 2226–2236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silver, P.A.; Way, J.C. Eukaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell 1993, 74, 5–6. [Google Scholar] [CrossRef]
- Lo, J.F.; Zhou, H.; Fearns, C.; Reisfeld, R.A.; Yang, Y.; Lee, J.D. Tid1 is required for T cell transition from double-negative 3 to double-positive stages. J. Immunol. 2005, 174, 6105–6112. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.H.; Hung, K.F.; Lee, T.C.; Huang, C.Y.; Chiu, W.T.; Lo, J.F.; Huang, T.F. Mitochondrial co-chaperone protein Tid1 is required for energy homeostasis during skeletal myogenesis. Stem. Cell Res. Ther. 2016, 7, 185. [Google Scholar] [CrossRef] [Green Version]
- Syken, J.; De-Medina, T.; Münger, K. TID1, a human homolog of the Drosophila tumor suppressor l(2)tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc. Natl. Acad. Sci. USA 1999, 96, 8499–8504. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.Y.; Jan, C.I.; Lo, J.F.; Yang, S.C.; Chang, Y.L.; Pan, S.H.; Wang, W.L.; Hong, T.M.; Yang, P.C. Tid1-L inhibits EGFR signaling in lung adenocarcinoma by enhancing EGFR Ubiquitinylation and degradation. Cancer Res. 2013, 73, 4009–4019. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.S.; Chang, C.W.; Tsay, Y.G.; Huang, L.Y.; Wu, Y.C.; Cheng, L.H.; Yang, C.C.; Wu, C.H.; Teo, W.H.; Hung, K.F.; et al. HSP40 co-chaperone protein Tid1 suppresses metastasis of head and neck cancer by inhibiting Galectin-7-TCF3-MMP9 axis signaling. Theranostics 2018, 8, 3841–3855. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chiou, S.H.; Huang, C.Y.; Jan, C.I.; Lin, S.C.; Hu, W.Y.; Chou, S.H.; Liu, C.J.; Lo, J.F. Tid1 functions as a tumour suppressor in head and neck squamous cell carcinoma. J. Pathol. 2009, 219, 347–355. [Google Scholar] [CrossRef]
- Wang, S.F.; Huang, K.H.; Tseng, W.C.; Lo, J.F.; Li, A.F.; Fang, W.L.; Chen, C.F.; Yeh, T.S.; Chang, Y.L.; Chou, Y.C.; et al. DNAJA3/Tid1 Is Required for Mitochondrial DNA Maintenance and Regulates Migration and Invasion of Human Gastric Cancer Cells. Cancers 2020, 12, 463. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.C.; Baird, S.D.; Screaton, R.A. Essential role of TID1 in maintaining mitochondrial membrane potential homogeneity and mitochondrial DNA integrity. Mol. Cell Biol. 2014, 34, 1427–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, B.; Ahn, Y.; Kang, S.M.; Park, Y.; Jeon, Y.J.; Rho, J.M.; Kim, S.W. Stoichiometric expression of mtHsp40 and mtHsp70 modulates mitochondrial morphology and cristae structure via Opa1L cleavage. Mol. Biol. Cell 2015, 26, 2156–2167. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Taslima, F.; Abdelhamid, M.; Kim, S.W.; Akatsu, H.; Michikawa, M.; Jung, C.G. Beta-Amyloid Increases the Expression Levels of Tid1 Responsible for Neuronal Cell Death and Amyloid Beta Production. Mol. Neurobiol. 2020, 57, 1099–1114. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tan, M.; Cai, Q. The Warburg effect in tumor progression: Mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015, 356, 156–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taguchi, K.; Motohashi, H.; Yamamoto, M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells 2011, 16, 123–140. [Google Scholar] [CrossRef]
- Xu, D.; Xu, M.; Jeong, S.; Qian, Y.; Wu, H.; Xia, Q.; Kong, X. The Role of Nrf2 in Liver Disease: Novel Molecular Mechanisms and Therapeutic Approaches. Front Pharmacol. 2018, 9, 1428. [Google Scholar] [CrossRef] [Green Version]
- Karin, M.; Dhar, D. Liver carcinogenesis: From naughty chemicals to soothing fat and the surprising role of NRF2. Carcinogenesis 2016, 37, 541–546. [Google Scholar] [CrossRef] [Green Version]
- Mitsuishi, Y.; Taguchi, K.; Kawatani, Y.; Shibata, T.; Nukiwa, T.; Aburatani, H.; Yamamoto, M.; Motohashi, H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 2012, 22, 66–79. [Google Scholar] [CrossRef] [Green Version]
- Zhan, L.; Zhang, H.; Zhang, Q.; Woods, C.G.; Chen, Y.; Xue, P.; Dong, J.; Tokar, E.J.; Xu, Y.; Hou, Y.; et al. Regulatory role of KEAP1 and NRF2 in PPARγ expression and chemoresistance in human non-small-cell lung carcinoma cells. Free Radic. Biol. Med. 2012, 53, 758–768. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Zhang, C.; Zhang, L.; Yang, Q.; Zhou, S.; Wen, Q.; Wang, J. Nrf2 is a potential prognostic marker and promotes proliferation and invasion in human hepatocellular carcinoma. BMC Cancer 2015, 15, 531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Ye, W.; Duan, X.; Zhang, M.; Wang, J. The noncytotoxic dose of sorafenib sensitizes Bel-7402/5-FU cells to 5-FU by down-regulating 5-FU-induced Nrf2 expression. Dig. Dis. Sci. 2013, 58, 1615–1626. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.W.; Chen, Y.S.; Tsay, Y.G.; Han, C.L.; Chen, Y.J.; Yang, C.C.; Hung, K.F.; Lin, C.H.; Huang, T.Y.; Kao, S.Y.; et al. ROS-independent ER stress-mediated NRF2 activation promotes warburg effect to maintain stemness-associated properties of cancer-initiating cells. Cell Death Dis. 2018, 9, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schilling, B.; De-Medina, T.; Syken, J.; Vidal, M.; Münger, K. A novel human DnaJ protein, hTid-1, a homolog of the Drosophila tumor suppressor protein Tid56, can interact with the human papillomavirus type 16 E7 oncoprotein. Virology 1998, 247, 74–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, H.; Cenciarelli, C.; Shao, Z.; Vidal, M.; Parks, W.P.; Pagano, M.; Cheng-Mayer, C. Human T cell leukemia virus type 1 Tax associates with a molecular chaperone complex containing hTid-1 and Hsp70. Curr. Biol. 2001, 11, 1771–1775. [Google Scholar] [CrossRef] [Green Version]
- Sohn, S.Y.; Kim, S.B.; Kim, J.; Ahn, B.Y. Negative regulation of hepatitis B virus replication by cellular Hsp40/DnaJ proteins through destabilization of viral core and X proteins. J. Gen. Virol. 2006, 87, 1883–1891. [Google Scholar] [CrossRef]
- Wang, L.; Tam, J.P.; Liu, D.X. Biochemical and functional characterization of Epstein-Barr virus-encoded BARF1 protein: Interaction with human hTid1 protein facilitates its maturation and secretion. Oncogene 2006, 25, 4320–4331. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.W.; Chao, T.H.; Xiang, R.; Lo, J.F.; Campbell, M.J.; Fearns, C.; Lee, J.D. Tid1, the human homologue of a Drosophila tumor suppressor, reduces the malignant activity of ErbB-2 in carcinoma cells. Cancer Res. 2004, 64, 7732–7739. [Google Scholar] [CrossRef] [Green Version]
- Koch, H.B.; Zhang, R.; Verdoodt, B.; Bailey, A.; Zhang, C.D.; Yates, J.R., 3rd; Menssen, A.; Hermeking, H. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle 2007, 6, 205–217. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yuan, Y.; Zhou, Y.; Guo, L.; Zhang, L.; Kuai, X.; Deng, B.; Pan, Z.; Li, D.; He, F. Protein interaction data set highlighted with human Ras-MAPK/PI3K signaling pathways. J. Proteome Res. 2008, 7, 3879–3889. [Google Scholar] [CrossRef]
- Bae, M.K.; Jeong, J.W.; Kim, S.H.; Kim, S.Y.; Kang, H.J.; Kim, D.M.; Bae, S.K.; Yun, I.; Trentin, G.A.; Rozakis-Adcock, M.; et al. Tid-1 interacts with the von Hippel-Lindau protein and modulates angiogenesis by destabilization of HIF-1alpha. Cancer Res. 2005, 65, 2520–2525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jan, C.I.; Yu, C.C.; Hung, M.C.; Harn, H.J.; Nieh, S.; Lee, H.S.; Lou, M.A.; Wu, Y.C.; Chen, C.Y.; Huang, C.Y.; et al. Tid1, CHIP and ErbB2 interactions and their prognostic implications for breast cancer patients. J. Pathol. 2011, 225, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Ahn, B.Y.; Trinh, D.L.; Zajchowski, L.D.; Lee, B.; Elwi, A.N.; Kim, S.W. Tid1 is a new regulator of p53 mitochondrial translocation and apoptosis in cancer. Oncogene 2010, 29, 1155–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trinh, D.L.; Elwi, A.N.; Kim, S.W. Direct interaction between p53 and Tid1 proteins affects p53 mitochondrial localization and apoptosis. Oncotarget 2010, 1, 396–404. [Google Scholar] [CrossRef]
- Teramoto, T.; Satonaka, K.; Kitazawa, S.; Fujimori, T.; Hayashi, K.; Maeda, S. p53 gene abnormalities are closely related to hepatoviral infections and occur at a late stage of hepatocarcinogenesis. Cancer Res. 1994, 54, 231–235. [Google Scholar]
- Surh, Y.J.; Kundu, J.K.; Li, M.H.; Na, H.K.; Cha, Y.N. Role of Nrf2-mediated heme oxygenase-1 upregulation in adaptive survival response to nitrosative stress. Arch. Pharm. Res. 2009, 32, 1163–1176. [Google Scholar] [CrossRef]
- Vasiliou, V.; Qamar, L.; Pappa, A.; Sophos, N.A.; Petersen, D.R. Involvement of the electrophile responsive element and p53 in the activation of hepatic stellate cells as a response to electrophile menadione. Arch. Biochem. Biophys. 2003, 413, 164–171. [Google Scholar] [CrossRef]
- Yeligar, S.M.; Machida, K.; Kalra, V.K. Ethanol-induced HO-1 and NQO1 are differentially regulated by HIF-1alpha and Nrf2 to attenuate inflammatory cytokine expression. J. Biol. Chem. 2010, 285, 35359–35373. [Google Scholar] [CrossRef] [Green Version]
- Ngo, H.K.C.; Kim, D.H.; Cha, Y.N.; Na, H.K.; Surh, Y.J. Nrf2 Mutagenic Activation Drives Hepatocarcinogenesis. Cancer Res. 2017, 77, 4797–4808. [Google Scholar] [CrossRef] [Green Version]
- Aydin, Y.; Chedid, M.; Chava, S.; Danielle Williams, D.; Liu, S.; Hagedorn, C.H.; Sumitran-Holgersson, S.; Reiss, K.; Moroz, K.; Lu, H.; et al. Activation of PERK-Nrf2 oncogenic signaling promotes Mdm2-mediated Rb degradation in persistently infected HCV culture. Sci. Rep. 2017, 7, 9223. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.; Wang, J.; Ji, X.; Cao, H.; Zhu, J.; Chen, Y.; Yang, J.; Zhao, Z.; Ren, T.; Xing, J. MCUR1 facilitates epithelial-mesenchymal transition and metastasis via the mitochondrial calcium dependent ROS/Nrf2/Notch pathway in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, M.C.; Hasegawa, K.; Chen, X.P.; Nagano, H.; Lee, Y.J.; Chau, G.Y.; Zhou, J.; Wang, C.C.; Choi, Y.R.; Poon, R.T.; et al. Surgery for Intermediate and Advanced Hepatocellular Carcinoma: A Consensus Report from the 5th Asia-Pacific Primary Liver Cancer Expert Meeting (APPLE 2014). Liver Cancer 2016, 5, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Imamura, H.; Matsuyama, Y.; Tanaka, E.; Ohkubo, T.; Hasegawa, K.; Miyagawa, S.; Sugawara, Y.; Minagawa, M.; Takayama, T.; Kawasaki, S.; et al. Risk factors contributing to early and late phase intrahepatic recurrence of hepatocellular carcinoma after hepatectomy. J. Hepatol. 2003, 38, 200–207. [Google Scholar] [CrossRef]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.F.; Chang, I.C.; Hong, C.C.; Yen, T.C.; Chen, C.L.; Wu, C.C.; Tsai, C.C.; Ho, M.C.; Lee, W.C.; Yu, H.C.; et al. Metabolic risk factors are associated with non-hepatitis B non-hepatitis C hepatocellular carcinoma in Taiwan, an endemic area of chronic hepatitis B. Hepatol. Commun. 2018, 2, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Hofman, V.; Rouquette, I.; Long-Mira, E.; Piton, N.; Chamorey, E.; Heeke, S.; Vignaud, J.M.; Yguel, C.; Mazieres, J.; Lepage, A.L.; et al. Multicenter Evaluation of a Novel ROS1 Immunohistochemistry Assay (SP384) for Detection of ROS1 Rearrangements in a Large Cohort of Lung Adenocarcinoma Patients. J. Thorac. Oncol. 2019, 14, 1204–1212. [Google Scholar] [CrossRef]





| Variables | HCC Patients (n = 210) |
|---|---|
| Age, years, mean ± SD | 59.23 ± 13.12 |
| Male sex, n (%) | 156 (74.3) |
| Drinking, n (%) | 67 (31.9) |
| Dual HBV and HCV infection, n (%) | 2 (1.0) |
| Alpha-fetoprotein, ng/mL, mean ± SD | 14,833.18 ± 64,194.02 |
| Tumor size, cm, mean ± SD | 6.15 ± 4.10 |
| Tumor number, n (%) | |
| n = 1/n > 1/Diffuse or infiltrative | 141/68/1 (67.1/32.4/0.5) |
| Cirrhosis, n (%) | 86 (41.0) |
| Differentiation, n (%) | |
| Well/Moderately/Poorly | 14/147/49 (6.7/70.0/23.3) |
| BCLC stage, n (%) | |
| A/B/C-D | 73/17/120 (34.8/8.1/57.1) |
| TNM stage, n (%) | |
| T1/T2/T3 | 73/24/113 (34.8/11.4/53.8) |
| T Stage | T1 + T2 | T3 | ||||
|---|---|---|---|---|---|---|
| H Score | Tumor | Non-Tumor | p Value | Tumor | Non-Tumor | p Value |
| HCC (all) * | 10 (0–165) | 55 (0–285) | <0.001 | 20 (0–185) | 85 (0–255) | <0.001 |
| HBV-HCC ** | 0 (0–165) | 57.5 (0–285) | <0.001 | 0 (0–185) | 97.5 (0–255) | <0.001 |
| HCV-HCC *** | 30 (0–130) | 55 (0–185) | 0.028 | 55 (0–145) | 70 (13–160) | 0.004 |
| TNM-T Stage/HCC Differentiation | Low H Score ≤ 75, n (%) | High H Score > 75, n (%) | p Value |
|---|---|---|---|
| HCC, n = 210 | |||
| TNM-T stage | 0.002 | ||
| T1 and T2, n = 93 | 63 (56.3) | 30 (33.3) | |
| T3, n = 109 | 49 (43.8) | 60 (66.7) | |
| Differentiation | 0.395 | ||
| Well/Moderate, n = 157 | 90 (80.4) | 67 (74.4) | |
| Poorly, n = 45 | 22 (19.6) | 23 (25.6) | |
| HBV-HCC, n = 105 | |||
| TNM-T stage | 0.017 | ||
| T1 and T2, n = 42 | 26 (53.1) | 16 (29.1) | |
| T3, n = 62 | 23 (46.9) | 39 (70.9) | |
| Differentiation | 0.426 | ||
| Well/Moderate, n = 87 | 43 (87.8) | 44 (80.0) | |
| Poorly, n = 17 | 6 (12.2) | 11 (20.0) | |
| HCV-HCC, n = 105 | |||
| TNM-T stage | 0.093 | ||
| T1 and T2, n = 51 | 37 (58.7) | 14 (40.0) | |
| T3, n = 47 | 26 (41.3) | 21 (60.0) | |
| Differentiation | 0.361 | ||
| Well/Moderate, n = 70 | 47 (74.6) | 23 (65.7) | |
| Poorly, n = 28 | 16 (25.4) | 12 (34.3) |
| TNM-T Stage/HCC Differentiation | Low H Score ≤ 47.25 | High H Score > 47.25 | p Value |
|---|---|---|---|
| HCC, n = 210 | |||
| TNM-T stage | 0.096 | ||
| T1 and T2, n = 95 | 72 (50.0) | 23 (37.1) | |
| T3, n = 111 | 72 (50.0) | 39 (62.9) | |
| Differentiation | 0.109 | ||
| Good/Moderate, n = 158 | 115 (79.9) | 43 (69.4) | |
| Poorly, n = 48 | 29 (20.1) | 19 (30.6) | |
| HBV-HCC, n = 105 | |||
| TNM-T stage | 0.197 | ||
| T1 and T2, n = 42 | 40 (42.6) | 2 (20.0) | |
| T3, n = 62 | 54 (57.4) | 8 (80.0) | |
| Differentiation | 0.361 | ||
| Good/Moderate, n = 87 | 77 (81.9) | 10 (100.0) | |
| Poorly, n = 17 | 17 (18.1) | 0 | |
| HCV-HCC, n = 105 | |||
| TNM-T stage | 0.019 | ||
| T1 and T2, n = 53 | 32 (64.0) | 21 (40.4) | |
| T3, n = 49 | 18 (36.0) | 31 (59.6) | |
| Differentiation | 0.200 | ||
| Good/Moderate, n = 71 | 38 (76.0) | 33 (63.5) | |
| Poorly, n = 31 | 12 (24.0) | 19 (36.5) |
| H Score, Median (Range) | Cirrhosis | Non-Cirrhosis | p Value |
|---|---|---|---|
| HCC * | 60 (0–210) | 80 (0–285) | <0.001 |
| HBV-HCC ** | 60 (0–210) (n = 41) | 85 (0–285) (n = 63) | 0.016 |
| HCV-HCC *** | 58 (0–108) (n = 43) | 70 (5–185) (n = 55) | 0.006 |
| Characteristics | Univariate | Multivariate I | Multivariate II | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | ||
| Age (years) | >50 vs. ≤50 | 1.080 | 0.726–1.607 | 0.703 | NA | NA | ||||
| Sex | Male vs. Female | 0.902 | 0.601–1.353 | 0.617 | NA | NA | ||||
| Alpha-fetoprotein (ng/mL) | ≥400 vs. <400 | 1.556 | 1.074–2.255 | 0.019 | NS | NS | ||||
| Liver Cirrhosis | Presence vs. Absence | 1.285 | 0.905–1.825 | 0.161 | NS | NS | ||||
| Underlying Hepatitis | HCV vs. HBV | 1.128 | 0.795–1.600 | 0.499 | NA | NA | ||||
| Tumor size (cm) | >5 vs. ≤5 | 1.573 | 1.109–2.230 | 0.011 | 1.618 | 1.074–2.439 | 0.021 | NA | ||
| Tumor number | >1 vs. ≤1 | 2.111 | 1.474–3.024 | <0.001 | 1.606 | 1.071–2.408 | 0.022 | NA | ||
| Vascular invasion | Presence vs. Absence | 1.798 | 1.268–2.552 | 0.001 | 1.658 | 1.104–2.490 | 0.015 | NA | ||
| TNM-T stage | T1 | 1.000 | - | NA | 1.000 | - | ||||
| T2 | 2.085 | 1.196–3.635 | 0.010 | NS | ||||||
| T3 | 2.582 | 1.700–3.922 | <0.001 | 2.508 | 1.593–3.949 | <0.001 | ||||
| TREND | <0.001 | <0.001 | ||||||||
| Pathology stage | 3–4 vs. 1–2 | 2.574 | 1.788–3.704 | <0.001 | NA | NA | ||||
| Differentiation | Good | 1.000 | - | NA | NA | |||||
| Moderate | 1.549 | 0.717–3.348 | 0.265 | |||||||
| Poor | 1.539 | 0.677–3.499 | 0.303 | |||||||
| TREND | 0.534 | |||||||||
| IHC expression, H score | ||||||||||
| Tid1, non-tumor part | >75 vs. ≤75 | 0.699 | 0.486–1.007 | 0.054 | 0.500 | 0.330–0.756 | 0.001 | 0.510 | 0.337–0.771 | 0.001 |
| Tid1, tumor part | >47.25 vs. ≤47.25 | 1.397 | 0.960–2.033 | 0.081 | NS | NS | ||||
| Nrf2-nucleus, non-tumor part | >5 vs. ≤5 | 1.082 | 0.663–1.763 | 0.753 | NA | NA | ||||
| Nrf2-nucleus, tumor part | >40 vs. ≤40 | 1.244 | 0.868–1.783 | 0.235 | NA | NA | ||||
| Nrf2-cytoplasma, non-tumor part | >82.5 vs. ≤82.5 | 1.421 | 0.999–2.021 | 0.050 | 1.527 | 1.023–2.280 | 0.038 | 1.531 | 1.027–2.282 | 0.036 |
| Nrf2-cytoplasma, tumor part | >95 vs. ≤95 | 1.490 | 1.049–2.116 | 0.026 | NS | NS | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, K.-Y.; Huang, Y.-H.; Teo, W.-H.; Chang, C.-W.; Chen, Y.-S.; Yeh, Y.-C.; Lee, C.-J.; Lo, J.-F. Loss of Tid1/DNAJA3 Co-Chaperone Promotes Progression and Recurrence of Hepatocellular Carcinoma after Surgical Resection: A Novel Model to Stratify Risk of Recurrence. Cancers 2021, 13, 138. https://doi.org/10.3390/cancers13010138
Chen K-Y, Huang Y-H, Teo W-H, Chang C-W, Chen Y-S, Yeh Y-C, Lee C-J, Lo J-F. Loss of Tid1/DNAJA3 Co-Chaperone Promotes Progression and Recurrence of Hepatocellular Carcinoma after Surgical Resection: A Novel Model to Stratify Risk of Recurrence. Cancers. 2021; 13(1):138. https://doi.org/10.3390/cancers13010138
Chicago/Turabian StyleChen, Kuan-Yang, Yi-Hsiang Huang, Wan-Huai Teo, Ching-Wen Chang, Yu-Syuan Chen, Yi-Chen Yeh, Chieh-Ju Lee, and Jeng-Fan Lo. 2021. "Loss of Tid1/DNAJA3 Co-Chaperone Promotes Progression and Recurrence of Hepatocellular Carcinoma after Surgical Resection: A Novel Model to Stratify Risk of Recurrence" Cancers 13, no. 1: 138. https://doi.org/10.3390/cancers13010138
APA StyleChen, K. -Y., Huang, Y. -H., Teo, W. -H., Chang, C. -W., Chen, Y. -S., Yeh, Y. -C., Lee, C. -J., & Lo, J. -F. (2021). Loss of Tid1/DNAJA3 Co-Chaperone Promotes Progression and Recurrence of Hepatocellular Carcinoma after Surgical Resection: A Novel Model to Stratify Risk of Recurrence. Cancers, 13(1), 138. https://doi.org/10.3390/cancers13010138