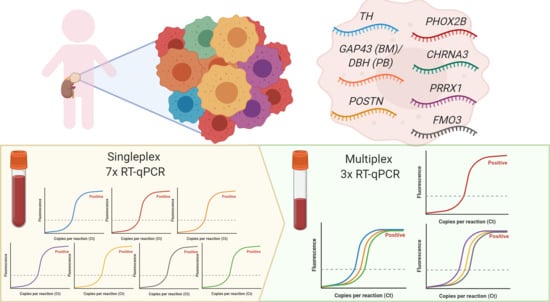
Specific and Sensitive Detection of Neuroblastoma mRNA Markers by Multiplex RT-qPCR
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Optimization of RT-qPCR Setting
2.2. Expression of Singleplex vs. MPX on Neuroblastoma Cell Lines
2.3. TH in MPX RT-qPCR Occasionally Showed Amplification of gDNA
2.4. Setting a Threshold for Positivity for ADRN and MES mRNA MPX in BM and PB
2.5. Comparison of Singleplex vs. MPX RT-qPCR in Patient Samples
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. RNA Isolation and cDNA Synthesis
4.3. Multiplex Real-Time qPCR and Sequencing
4.4. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADRN | adrenergic (neuroblastoma) |
| BHQ | Black Hole Quencher |
| Ct | cycle threshold |
| DFO | Dragonfly Orange™ |
| dNTP | deoxyribonucleotide triphosphate |
| DTT | dithiothreitol |
| EMT | epithelial to mesenchymal transition |
| MES | mesenchymal (neuroblastoma) |
| MMLV | Moloney Murine Leukemia Virus |
| MPX | multiplex RT-qPCR |
| NBL | neuroblastoma |
| PB | peripheral blood |
| YY | Yakima Yellow™ |
Genes
| CHRNA3 | cholinergic receptor nicotinic alpha 3 |
| DBH | dopamine beta hydroxylase |
| FMO3 | flavin containing monooxygenase 3 |
| GAP43 | growth associated protein 43 |
| GUSB | glucuronidase beta |
| PHOX2B | paired-like homeobox 2b |
| POSTN | periostin |
| PRRX1 | paired related homeobox 1 |
| TH | tyrosine hydroxylase |
References
- Cohn, S.L.; Pearson, A.D.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D. The international neuroblastoma risk group (inrg) classification system: An inrg task force report. J. Clin. Oncol. 2009, 27, e289. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; Villablanca, J.G.; Seeger, R.C.; Stram, D.O.; Harris, R.E.; Ramsay, N.K.; Swift, P.; Shimada, H.; Black, C.T.; Brodeur, G.M. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. N. Engl. J. Med. 1999, 341, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Viprey, V.F.; Gregory, W.M.; Corrias, M.V.; Tchirkov, A.; Swerts, K.; Vicha, A.; Dallorso, S.; Brock, P.; Luksch, R.; Valteau-Couanet, D.; et al. Neuroblastoma mrnas predict outcome in children with stage 4 neuroblastoma: A european hr-nbl1/siopen study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Stutterheim, J.; Zappeij-Kannegieter, L.; Versteeg, R.; Caron, H.N.; van der Schoot, C.E.; Tytgat, G.A. The prognostic value of fast molecular response of marrow disease in patients aged over 1 year with stage 4 neuroblastoma. Eur. J. Cancer 2011, 47, 1193–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, J.-Y.; Pan, C.; Tang, Y.-J.; Chen, J.; Ye, Q.-D.; Zhou, M.; Xue, H.; Tang, J.-Y. Minimal residual disease is a prognostic marker for neuroblastoma with bone marrow infiltration. Am. J. Clin. Oncol. 2012, 35, 275–278. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.B.; Bae, G.E.; Lee, N.H.; Kim, J.-S.; Lee, S.H.; Yoo, K.H.; Sung, K.W.; Koo, H.H. Clinical significance of persistent tumor in bone marrow during treatment of high-risk neuroblastoma. J. Korean Med Sci. 2015, 30, 1062–1067. [Google Scholar] [CrossRef]
- Seeger, R.C.; Reynolds, C.P.; Gallego, R.; Stram, D.O.; Gerbing, R.B.; Matthay, K.K. Quantitative tumor cell content of bone marrow and blood as a predictor of outcome in stage iv neuroblastoma: A children’s cancer group study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2000, 18, 4067–4076. [Google Scholar] [CrossRef]
- Stutterheim, J.; Gerritsen, A.; Zappeij-Kannegieter, L.; Kleijn, I.; Dee, R.; Hooft, L.; van Noesel, M.M.; Bierings, M.; Berthold, F.; Versteeg, R.; et al. Phox2b is a novel and specific marker for minimal residual disease testing in neuroblastoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2008, 26, 5443–5449. [Google Scholar] [CrossRef]
- Stutterheim, J.; Gerritsen, A.; Zappeij-Kannegieter, L.; Yalcin, B.; Dee, R.; van Noesel, M.M.; Berthold, F.; Versteeg, R.; Caron, H.N.; van der Schoot, C.E.; et al. Detecting minimal residual disease in neuroblastoma: The superiority of a panel of real-time quantitative PCR markers. Clin. Chem. 2009, 55, 1316–1326. [Google Scholar] [CrossRef] [Green Version]
- Beiske, K.; Burchill, S.A.; Cheung, I.Y.; Hiyama, E.; Seeger, R.C.; Cohn, S.L.; Pearson, A.D.; Matthay, K.K.; International neuroblastoma Risk Group Task, F. Consensus criteria for sensitive detection of minimal neuroblastoma cells in bone marrow, blood and stem cell preparations by immunocytology and qrt-pcr: Recommendations by the international neuroblastoma risk group task force. Br. J. Cancer 2009, 100, 1627–1637. [Google Scholar] [CrossRef]
- Burchill, S.A.; Lewis, I.J.; Abrams, K.R.; Riley, R.; Imeson, J.; Pearson, A.D.; Pinkerton, R.; Selby, P. Circulating neuroblastoma cells detected by reverse transcriptase polymerase chain reaction for tyrosine hydroxylase mrna are an independent poor prognostic indicator in stage 4 neuroblastoma in children over 1 year. J. Clin. Oncol. 2001, 19, 1795–1801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tchirkov, A.; Paillard, C.; Halle, P.; Bernard, F.; Bordigoni, P.; Vago, P.; Deméocq, F.; Kanold, J. Significance of molecular quantification of minimal residual disease in metastatic neuroblastoma. J. Hematotherapy Stem Cell Res. 2003, 12, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Wezel, E.M.V.; Zogchel, L.M.J.V.; Wijk, J.V.; Timmerman, I.; Vo, N.-K.; Zappeij-Kannegieter, L.; de Carolis, B.; Simon, T.; Noesel, M.M.V.; Molenaar, J.J.; et al. Mesenchymal neuroblastoma cells are undetected by current mrna marker panels: The development of a specific neuroblastoma mesenchymal minimal residual disease panel. JCO Precis. Oncol. 2019, 3, 1–11. [Google Scholar]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. Emt in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef]
- van Groningen, T.; Koster, J.; Valentijn, L.J.; Zwijnenburg, D.A.; Akogul, N.; Hasselt, N.E.; Broekmans, M.; Haneveld, F.; Nowakowska, N.E.; Bras, J.; et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 2017, 49, 1261–1266. [Google Scholar] [CrossRef]
- Boiso, L.; Sanga, M.; Hedman, J. Dtt quenches the passive reference signal in real-time PCR. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e5–e6. [Google Scholar] [CrossRef]
- Burchill, S.A.; Beiske, K.; Shimada, H.; Ambros, P.F.; Seeger, R.; Tytgat, G.A.; Brock, P.R.; Haber, M.; Park, J.R.; Berthold, F. Recommendations for the standardization of bone marrow disease assessment and reporting in children with neuroblastoma on behalf of the international neuroblastoma response criteria bone marrow working group. Cancer 2017, 123, 1095–1105. [Google Scholar] [CrossRef]
- Viprey, V.F.; Lastowska, M.A.; Corrias, M.V.; Swerts, K.; Jackson, M.S.; Burchill, S.A. Minimal disease monitoring by qrt-pcr: Guidelines for identification and systematic validation of molecular markers prior to evaluation in prospective clinical trials. J. Pathol. 2008, 216, 245–252. [Google Scholar] [CrossRef]
- Applied biosystems™ Relative Quantitation Analysis Module User Guide. C.0 ed. ThermoFisher Scientific, 2016. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/MAN0014820_RelativeQuant_UG.pdf (accessed on 9 June 2020).
- van der Velden, V.H.; Cazzaniga, G.; Schrauder, A.; Hancock, J.; Bader, P.; Panzer-Grumayer, E.R.; Flohr, T.; Sutton, R.; Cave, H.; Madsen, H.O.; et al. Analysis of minimal residual disease by ig/tcr gene rearrangements: Guidelines for interpretation of real-time quantitative PCR data. Leukemia 2007, 21, 604–611. [Google Scholar] [CrossRef] [Green Version]
- Stutterheim, J.; Zappeij-Kannegieter, L.; Ora, I.; van Sluis, P.G.; Bras, J.; den Ouden, E.; Versteeg, R.; Caron, H.N.; van der Schoot, C.E.; Tytgat, G.A. Stability of PCR targets for monitoring minimal residual disease in neuroblastoma. J. Mol. Diagn. Jmd 2012, 14, 168–175. [Google Scholar] [CrossRef]
- Boeva, V.; Louis-Brennetot, C.; Peltier, A.; Durand, S.; Pierre-Eugene, C.; Raynal, V.; Etchevers, H.C.; Thomas, S.; Lermine, A.; Daudigeos-Dubus, E. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat. Genet. 2017, 49, e1408. [Google Scholar] [CrossRef] [PubMed]
- Bethesda (MD): National Library of Medicine (US), N.C.f.B.I. Available online: https://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&id=1675122364 (accessed on 30 September 2020).
- Ucsc Genome Browser on Human Dec. 2013 (grch38/hg38) Assembly. Available online: https://genome-euro.ucsc.edu/cgi-bin/hgTracks?db=hg38&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr1%3A171090905%2D171117819&hgsid=245864441_abIEEt4GrIAlPZztyxlE9hklBgQa (accessed on 30 October 2020).
- Debode, F.; Marien, A.; Janssen, É.; Bragard, C.; Berben, G. The influence of amplicon length on real-time PCR results. Base 2017. [Google Scholar] [CrossRef]
- Beillard, E.; Pallisgaard, N.; van der Velden, V.H.; Bi, W.; Dee, R.; van der Schoot, E.; Delabesse, E.; Macintyre, E.; Gottardi, E.; Saglio, G.; et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (rq-pcr)-a europe against cancer program. Leukemia 2003, 17, 2474–2486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabert, J.; Beillard, E.; van der Velden, V.H.; Bi, W.; Grimwade, D.; Pallisgaard, N.; Barbany, G.; Cazzaniga, G.; Cayuela, J.M.; Cave, H.; et al. Standardization and quality control studies of ‘real-time’ quantitative reverse transcriptase polymerase chain reaction of fusion gene transcripts for residual disease detection in leukemia-a europe against cancer program. Leukemia 2003, 17, 2318–2357. [Google Scholar] [CrossRef]
- Simon, T.; Berthold, F.; Borkhardt, A.; Kremens, B.; De Carolis, B.; Hero, B. Treatment and outcomes of patients with relapsed, high-risk neuroblastoma: Results of german trials. Pediatric Blood Cancer 2011, 56, 578–583. [Google Scholar] [CrossRef]
- Kraal, K.C.; Bleeker, G.M.; van Eck-Smit, B.L.; van Eijkelenburg, N.K.; Berthold, F.; van Noesel, M.M.; Caron, H.N.; Tytgat, G.A. Feasibility, toxicity and response of upfront metaiodobenzylguanidine therapy therapy followed by german pediatric oncology group neuroblastoma 2004 protocol in newly diagnosed stage 4 neuroblastoma patients. Eur. J. Cancer 2017, 76, 188–196. [Google Scholar] [CrossRef]
- Bate-Eya, L.T.; Ebus, M.E.; Koster, J.; den Hartog, I.J.; Zwijnenburg, D.A.; Schild, L.; van der Ploeg, I.; Dolman, M.E.; Caron, H.N.; Versteeg, R.; et al. Newly-derived neuroblastoma cell lines propagated in serum-free media recapitulate the genotype and phenotype of primary neuroblastoma tumours. Eur. J. Cancer 2014, 50, 628–637. [Google Scholar] [CrossRef]
- van Nes, J.; Chan, A.; van Groningen, T.; van Sluis, P.; Koster, J.; Versteeg, R. A notch3 transcriptional module induces cell motility in neuroblastoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 3485–3494. [Google Scholar] [CrossRef] [Green Version]
- van Wezel, E.M.; Zwijnenburg, D.; Zappeij-Kannegieter, L.; Bus, E.; van Noesel, M.M.; Molenaar, J.J.; Versteeg, R.; Fiocco, M.; Caron, H.N.; van der Schoot, C.E.; et al. Whole-genome sequencing identifies patient-specific DNA minimal residual disease markers in neuroblastoma. J. Mol. Diagn. Jmd 2015, 17, 43–52. [Google Scholar] [CrossRef]
- Swerts, K.; De Moerloose, B.; Dhooge, C.; Vandesompele, J.; Hoyoux, C.; Beiske, K.; Benoit, Y.; Laureys, G.; Philippé, J. Potential application of elavl4 real-time quantitative reverse transcription-PCR for detection of disseminated neuroblastoma cells. Clin. Chem. 2006, 52, 438–445. [Google Scholar] [CrossRef] [Green Version]



| Marker | BM | PB | ||||
|---|---|---|---|---|---|---|
| Positive BM Samples | Expression (Median + IQR) | Threshold | Positive PB Samples | Expression (Median + IQR) | Threshold | |
| PHOX2B | 0/54 | 0/50 | ||||
| TH | 17/54 | 16.0 (0.9) | 13.0 | 21/50 | 14.3 (0.9) | 11.3 |
| CHRNA3 | 42/54 | 16.0 (1.7) | 13.0 | 21/50 | 16.4 (1.7) | 13.4 |
| GAP43 | 42/54 | 17.0 (2.3) | 14.0 | |||
| DBH | 6/50 | 16.7 (0.7) | 13.7 | |||
| FMO3 | 54/54 | 8.6 (2.7) | 16/50 | 15.2 (1.4) | ||
| PRRX1 | 54/54 | 10.3 (2.8) | 9.0 * | 30/50 | 15.3 (1.7) | 12.3 ** |
| POSTN | 54/54 | 12.1 (3.0) | 9.0 * | 27/50 | 15.4 (2.0) | 12.5 ** |
| Marker | BM (n = 24) | PB (n = 21) | ||
|---|---|---|---|---|
| MPX Positive BM Samples | Singpleplex Positive BM Samples | MPX Positive PB Samples | Singleplex Positive PB Samples | |
| PHOX2B | 24 | 24 | 20 | 19 |
| TH | 22 | 22 | 13 | 18 |
| CHRNA3 | 20 | 20 | 16 | 16 |
| GAP43 | 24 | 18 | ||
| DBH | 18 | 21 | ||
| Total ADRN | 24 * | 24 | 20 ** | 21 |
| PRRX1 | 9 | 9 | 0 | 0 |
| POSTN | 7 | 6 | 0 | 0 |
| Total MES | 11 *** | 10 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Zogchel, L.M.J.; Zappeij-Kannegieter, L.; Javadi, A.; Lugtigheid, M.; Gelineau, N.U.; Lak, N.S.M.; Zwijnenburg, D.A.; Koster, J.; Stutterheim, J.; van der Schoot, C.E.; et al. Specific and Sensitive Detection of Neuroblastoma mRNA Markers by Multiplex RT-qPCR. Cancers 2021, 13, 150. https://doi.org/10.3390/cancers13010150
van Zogchel LMJ, Zappeij-Kannegieter L, Javadi A, Lugtigheid M, Gelineau NU, Lak NSM, Zwijnenburg DA, Koster J, Stutterheim J, van der Schoot CE, et al. Specific and Sensitive Detection of Neuroblastoma mRNA Markers by Multiplex RT-qPCR. Cancers. 2021; 13(1):150. https://doi.org/10.3390/cancers13010150
Chicago/Turabian Stylevan Zogchel, Lieke M. J., Lily Zappeij-Kannegieter, Ahmad Javadi, Marjolein Lugtigheid, Nina U. Gelineau, Nathalie S. M. Lak, Danny A. Zwijnenburg, Jan Koster, Janine Stutterheim, C. Ellen van der Schoot, and et al. 2021. "Specific and Sensitive Detection of Neuroblastoma mRNA Markers by Multiplex RT-qPCR" Cancers 13, no. 1: 150. https://doi.org/10.3390/cancers13010150
APA Stylevan Zogchel, L. M. J., Zappeij-Kannegieter, L., Javadi, A., Lugtigheid, M., Gelineau, N. U., Lak, N. S. M., Zwijnenburg, D. A., Koster, J., Stutterheim, J., van der Schoot, C. E., & Tytgat, G. A. M. (2021). Specific and Sensitive Detection of Neuroblastoma mRNA Markers by Multiplex RT-qPCR. Cancers, 13(1), 150. https://doi.org/10.3390/cancers13010150