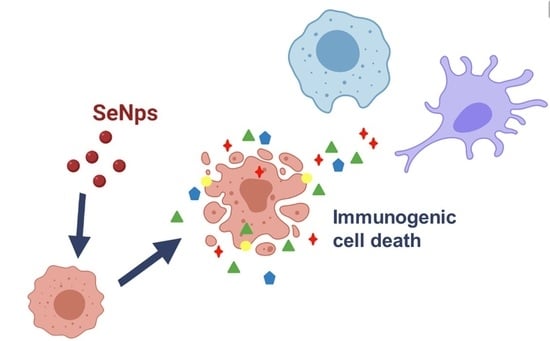
Anticancer Activity of Biogenic Selenium Nanoparticles: Apoptotic and Immunogenic Cell Death Markers in Colon Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell lines and Bacterial Cultures
2.3. Selenium Nanoparticles
2.4. TEM Analysis
2.5. Micro Fourier Transform Infrared Spectroscopy
2.6. Cell Growth Assay
2.7. Western Blot
2.8. Analysis of Apoptosis with Annexin V/PI Staining
2.9. Detection of Caspase 3 Clavage by Flow Cytometry
2.10. M30 Apoptosense Assay
2.11. Calreticulin and ERp57 Exposure Analysis by Flow Cytometry
2.12. Flow Cytometry Analysis of Intracellular ATP
2.13. Detection of Annexin V / PI Staining, HMGB1 Translocation, Caspase 3 Cleavage and ATP Depletion by Fluorescence Microscopy
2.14. Detection of Apoptosis Related Proteins in CT26 Cells with a Membrane-Based Sandwich Immunoassay
2.15. Quantification of Cytokines in SeNps-Treated Cells Supernatants
2.16. Assessment of Phagocytic Activity in Macrophages and SeNps-Treated Tumour Cells Co-Culture
2.17. Data Analysis and Statistics
3. Results
3.1. Characteristics of SeNps
3.2. Induction of Apoptosis in Colon Cancer Cells Treated with SeNps
3.2.1. SeNps Induce Apoptosis in Human Colon Cancer Cells
3.2.2. SeNps Induce Apoptosis in CT26 Cells
3.3. Induction of Immunogenic Cell Death in CT26 and HT29 Cells by SeNps
3.3.1. Release of Nuclear Protein HMGB1 from SeNps-Treated Cells
3.3.2. Surface Exposure of Calreticulin and ERp57 in SeNps-Treated Cells
3.3.3. SeNps Induce the Secretion of ATP from Cells
3.3.4. Secretion of Pro-Inflammatory Cytokines from SeNps-Treated Cells
3.4. Cancer CT26 Cells Destroyed by SeNps Are Preferentially Phagocytosed by RAW246.7 Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tapiero, H.; Townsend, D.M.; Tew, K.D. The antioxidant role of selenium and seleno-compounds. Biomed. Pharmacother. 2003, 57, 134–144. [Google Scholar] [CrossRef]
- Hosnedlova, B.; Kepinska, M.; Skalickova, S.; Fernandez, C.; Ruttkay-Nedecky, B.; Malevu, T.D.; Sochor, J.; Baron, M.; Melcova, M.; Zidkova, J.; et al. A Summary of New Findings on the Biological Effects of Selenium in Selected Animal Species—A Critical Review. Int. J. Mol. Sci. 2017, 18, 2209. [Google Scholar] [CrossRef]
- Hatfield, D.L.; Tsuji, P.A.; Carlson, B.A.; Gladyshev, V.N. Selenium and selenocysteine: Roles in cancer, health, and development. Trends Biochem. Sci. 2014, 39, 112–120. [Google Scholar] [CrossRef] [Green Version]
- Sanmartín, C.; Plano, D.; Sharma, A.K.; Palop, J.A. Selenium compounds, apoptosis and other types of cell death: An overview for cancer therapy. Int. J. Mol. Sci. 2012, 13, 9649–9672. [Google Scholar] [CrossRef]
- Vinceti, M.; Filippini, T.; Cilloni, S.; Crespi, C.M. The Epidemiology of Selenium and Human Cancer, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; Volume 136. [Google Scholar]
- Liu, M.; Yao, W.; Zhu, Y.; Liu, H.; Zhang, J.; Jia, L. Characterization, antioxidant and antiinflammation of mycelia selenium polysaccharides from Hypsizygus marmoreus SK-03. Carbohydr. Polym. 2018, 201, 566–574. [Google Scholar] [CrossRef]
- Khurana, A.; Tekula, S.; Saifi, M.A.; Venkatesh, P.; Godugu, C. Therapeutic applications of selenium nanoparticles. Biomed. Pharmacother. 2019, 111, 802–812. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, K.L. Evaluating selenium poisoning. Ann. Clin. Lab. Sci. 2006, 36, 409–420. [Google Scholar] [PubMed]
- Kuršvietienė, L.; Mongirdienė, A.; Bernatonienė, J.; Šulinskienė, J.; Stanevičienė, I. Selenium anticancer properties and impact on cellular redox status. Antioxidants 2020, 9, 80. [Google Scholar] [CrossRef] [Green Version]
- Hosnedlova, B.; Kepinska, M.; Skalickova, S.; Fernandez, C.; Ruttkay-Nedecky, B.; Peng, Q.; Baron, M.; Melcova, M.; Opatrilova, R.; Zidkova, J.; et al. Nano-selenium and its nanomedicine applications: A critical review. Int. J. Nanomed. 2018, 13, 2107–2128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maiyo, F.; Singh, M. Selenium nanoparticles: Potential in cancer gene and drug delivery. Nanomedicine 2017, 12, 1075–1089. [Google Scholar] [CrossRef]
- Ferro, C.; Florindo, H.F.; Santos, H.A. Selenium Nanoparticles for Biomedical Applications: From Development and Characterization to Therapeutics. Adv. Healthc. Mater. 2021, 2100598, 1–50. [Google Scholar]
- Husen, A.; Siddiqi, K.S. Phytosynthesis of nanoparticles: Concept, controversy and application. Nanoscale Res. Lett. 2014, 9, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tejo Prakash, N.; Sharma, N.; Prakash, R.; Raina, K.K.; Fellowes, J.; Pearce, C.I.; Lloyd, J.R.; Pattrick, R.A.D.D.; Prakash, N.T.; Sharma, N.; et al. Aerobic microbial manufacture of nanoscale selenium: Exploiting nature’s bio-nanomineralization potential. Biotechnol. Lett. 2009, 31, 1857–1862. [Google Scholar] [CrossRef] [PubMed]
- Tugarova, A.V.; Kamnev, A.A. Proteins in microbial synthesis of selenium nanoparticles. Talanta 2017, 174, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Oremland, R.S.; Herbel, M.J.; Blum, J.S.; Langley, S.; Beveridge, T.J.; Ajayan, P.M.; Sutto, T.; Ellis, A.V.; Curran, S. Structural and Spectral Features of Selenium Nanospheres Produced by Se-Respiring Bacteria. Appl. Environ. Microbiol. 2004, 70, 52–60. [Google Scholar] [CrossRef] [Green Version]
- Menon, S.; Ks, S.D.; Santhiya, R.; Rajeshkumar, S.; Kumar, V. Selenium nanoparticles: A potent chemotherapeutic agent and an elucidation of its mechanism. Colloids Surf. B. Biointerfaces 2018, 170, 280–292. [Google Scholar] [CrossRef]
- Gao, X.; Li, X.; Mu, J.; Ho, C.T.; Su, J.; Zhang, Y.; Lin, X.; Chen, Z.; Li, B.; Xie, Y. Preparation, physicochemical characterization, and anti-proliferation of selenium nanoparticles stabilized by Polyporus umbellatus polysaccharide. Int. J. Biol. Macromol. 2020, 152, 605–615. [Google Scholar] [CrossRef]
- Lin, W.; Zhang, J.; Xu, J.-F.; Pi, J. The Advancing of Selenium Nanoparticles Against Infectious Diseases. Front. Pharmacol. 2021, 12, 1971. [Google Scholar] [CrossRef]
- Deutsch, E.; Chargari, C.; Galluzzi, L.; Kroemer, G. Optimising efficacy and reducing toxicity of anticancer radioimmunotherapy. Lancet Oncol. 2019, 20, e452–e463. [Google Scholar] [CrossRef]
- Galluzzi, L.; Humeau, J.; Buqué, A.; Zitvogel, L.; Kroemer, G. Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2020, 17, 725–741. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I.; Warren, S.; Adjemian, S.; Agostinis, P.; Martinez, A.B.; Chan, T.A.; Coukos, G.; Demaria, S.; Deutsch, E.; et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J. Immunother. Cancer 2020, 8, e000337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yatim, N.; Cullen, S.; Albert, M.L. Dying cells actively regulate adaptive immune responses. Nat. Rev. Immunol. 2017, 17, 262–275. [Google Scholar] [CrossRef] [PubMed]
- Gong, T.; Liu, L.; Jiang, W.; Zhou, R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2020, 20, 95–112. [Google Scholar] [CrossRef] [PubMed]
- Fucikova, J.; Kepp, O.; Kasikova, L.; Petroni, G.; Yamazaki, T.; Liu, P.; Zhao, L.; Spisek, R.; Kroemer, G.; Galluzzi, L. Detection of immunogenic cell death and its relevance for cancer therapy. Cell Death Dis. 2020, 11, 1013. [Google Scholar] [CrossRef]
- Xiao, R.; Allen, C.T.; Tran, L.; Patel, P.; Park, S.J.; Chen, Z.; van Waes, C.; Schmitt, N.C. Antagonist of cIAP1/2 and XIAP enhances anti-tumor immunity when combined with radiation and PD-1 blockade in a syngeneic model of head and neck cancer. Oncoimmunology 2018, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Gunti, S.; Allen, C.T.; Hong, Y.; Clavijo, P.E.; van Waes, C.; Schmitt, N.C. ASTX660, an antagonist of cIAP1/2 and XIAP, increases antigen processing machinery and can enhance radiation-induced immunogenic cell death in preclinical models of head and neck cancer. Oncoimmunology 2020, 9, 1710398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, H.; Leibowitz, B.J.; Zhang, L.; Yu, J. Immunogenic cell death in colon cancer prevention and therapy. Mol. Carcinog. 2020, 59, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ju, X.; Wang, J.; Fan, Y.; Ren, M.; Zhang, H. Immunogenic cell death in anticancer chemotherapy and its impact on clinical studies. Cancer Lett. 2018, 438, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Pol, J.; Vacchelli, E.; Aranda, F.; Castoldi, F.; Eggermont, A.; Cremer, I.; Sautès-Fridman, C.; Fucikova, J.; Galon, J.; Spisek, R.; et al. Trial Watch: Immunogenic cell death inducers for anticancer chemotherapy. Oncoimmunology 2015, 4, e1008866. [Google Scholar] [CrossRef]
- D’Amico, L.; Menzel, U.; Prummer, M.; Müller, P.; Buchi, M.; Kashyap, A.; Haessler, U.; Yermanos, A.; Gébleux, R.; Briendl, M.; et al. A novel anti-HER2 anthracycline-based antibody-drug conjugate induces adaptive anti-tumor immunity and potentiates PD-1 blockade in breast cancer. J. Immunother. Cancer 2019, 7, 1–15. [Google Scholar] [CrossRef]
- Yamazaki, T.; Buqué, A.; Ames, T.D.; Galluzzi, L. PT-112 induces immunogenic cell death and synergizes with immune checkpoint blockers in mouse tumor models. Oncoimmunology 2020, 9, 1721810. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Zhao, L.; Pol, J.; Levesque, S.; Petrazzuolo, A.; Pfirschke, C.; Engblom, C.; Rickelt, S.; Yamazaki, T.; Iribarren, K.; et al. Crizotinib-induced immunogenic cell death in non-small cell lung cancer. Nat. Commun. 2019, 10, 1486. [Google Scholar] [CrossRef] [PubMed]
- Voorwerk, L.; Slagter, M.; Horlings, H.M.; Sikorska, K.; van de Vijver, K.K.; de Maaker, M.; Nederlof, I.; Kluin, R.J.C.; Warren, S.; Ong, S.F.; et al. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: The TONIC trial. Nat. Med. 2019, 25, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Antonia, S.J.; Villegas, A.; Daniel, D.; Vicente, D.; Murakami, S.; Hui, R.; Yokoi, T.; Chiappori, A.; Lee, K.H.; de Wit, M.; et al. Durvalumab after Chemoradiotherapy in Stage III Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 1919–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chang, Y.; Luo, H.; Jiang, W.; Xu, L.; Chen, T.; Zhu, X. Designing immunogenic nanotherapeutics for photothermal-triggered immunotherapy involving reprogramming immunosuppression and activating systemic antitumor responses. Biomaterials 2020, 255, 120153. [Google Scholar] [CrossRef] [PubMed]
- Yazdi, M.H.; Varastehmoradi, B.; Faghfuri, E.; Mavandadnejad, F.; Mahdavi, M.; Shahverdi, A.R. Adjuvant effect of biogenic selenium nanoparticles improves the immune responses and survival of mice receiving 4T1 cell antigens as vaccine in breast cancer murine model. J. Nanosci. Nanotechnol. 2015, 15, 10165–10172. [Google Scholar] [CrossRef]
- Gautam, P.K.; Kumar, S.; Tomar, M.S.; Singh, R.K.; Acharya, A.; Kumar, S.; Ram, B. Selenium nanoparticles induce suppressed function of tumor associated macrophages and inhibit Dalton’s lymphoma proliferation. Biochem. Biophys. Rep. 2017, 12, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Liu, T.; Li, J.J.; Mai, F.; Li, J.J.; Chen, Y.; Jing, Y.; Dong, X.; Lin, L.; He, J.; et al. Selenium nanoparticles as new strategy to potentiate γδ T cell anti-tumor cytotoxicity through upregulation of tubulin-α acetylation. Biomaterials 2019, 222, 119397. [Google Scholar] [CrossRef]
- Ryan-Harshman, M.; Aldoori, W. The relevance of selenium to immunity, cancer, and infectious/inflammatory diseases. Can. J. Diet. Pract. Res. 2005, 66, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Spyridopoulou, K.; Tryfonopoulou, E.; Aindelis, G.; Ypsilantis, P.; Sarafidis, C.; Kalogirou, O.; Chlichlia, K. Biogenic selenium nanoparticles produced by Lactobacillus casei ATCC 393 inhibit colon cancer cell growth in vitro and in vivo. Nanoscale Adv. 2021, 3, 2516–2528. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Kramer, G.; Erdal, H.; Mertens, H.J.M.M.; Nap, M.; Mauermann, J.; Steiner, G.; Marberger, M.; Bive, K.; Shoshan, M.C.; Linder, S. Differentiation between Cell Death Modes Using Measurements of Different Soluble Forms of Extracellular Cytokeratin 18. Cancer Res. 2004, 64, 1751–1756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kepp, O.; Tartour, E.; Vitale, I.; Vacchelli, E.; Adjemian, S.; Agostinis, P.; Apetoh, L.; Aranda, F.; Barnaba, V.; Bloy, N.; et al. Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology 2014, 3, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pangrsic, T.; Potokar, M.; Stenovec, M.; Kreft, M.; Fabbretti, E.; Nistri, A.; Pryazhnikov, E.; Khiroug, L.; Giniatullin, R.; Zorec, R. Exocytotic release of ATP from cultured astrocytes. J. Biol. Chem. 2007, 282, 28749–28758. [Google Scholar] [CrossRef] [Green Version]
- Forveille, S.; Humeau, J.; Sauvat, A.; Bezu, L.; Kroemer, G.; Kepp, O. Quinacrine-Mediated Detection of Intracellular ATP. In Methods Enzymol, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2019; Volume 629, ISBN 9780128186718. [Google Scholar]
- Jakic, B.; Buszko, M.; Cappellano, G.; Wick, G. Elevated sodium leads to the increased expression of HSP60 and induces apoptosis in HUVECs. PLoS ONE 2017, 12, e0179383. [Google Scholar] [CrossRef] [Green Version]
- McCloy, R.A.; Rogers, S.; Caldon, C.E.; Lorca, T.; Castro, A.; Burgess, A. Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle 2014, 13, 1400–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Qiao, L.; Ma, L.; Yan, S.; Guo, Y.; Dou, X.; Zhang, B.; Roman, A. Biosynthesis of Polysaccharides-Capped Selenium Nanoparticles Using Lactococcus lactis NZ9000 and Their Antioxidant and Anti-inflammatory Activities. Front. Microbiol. 2019, 10, 1632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Qiao, L.; Guo, Y.; Ma, L.; Cheng, Y. Preparation, characteristics and antioxidant activity of polysaccharides and proteins-capped selenium nanoparticles synthesized by Lactobacillus casei ATCC 393. Carbohydr. Polym. 2018, 195, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Kamnev, A.A.; Dyatlova, Y.A.; Kenzhegulov, O.A.; Vladimirova, A.A.; Mamchenkova, P.V.; Tugarova, A.V. Fourier Transform Infrared (FTIR) Spectroscopic Analyses of Microbiological Samples and Biogenic Selenium Nanoparticles of Microbial Origin: Sample Preparation Effects. Molecules 2021, 26, 1146. [Google Scholar] [CrossRef]
- Tugarova, A.V.; Mamchenkova, P.V.; Dyatlova, Y.A.; Kamnev, A.A. FTIR and Raman spectroscopic studies of selenium nanoparticles synthesised by the bacterium Azospirillum thiophilum. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 192, 458–463. [Google Scholar] [CrossRef]
- Wenning, M.; Scherer, S.; Naumann, D. Infrared Spectroscopy in the Identification of Microorganisms. Handb. Vib. Spectrosc. 2008, 1, 1–26. [Google Scholar]
- Spyridopoulou, K.; Tiptiri-Kourpeti, A.; Lampri, E.; Fitsiou, E.; Vasileiadis, S.; Vamvakias, M.; Bardouki, H.; Goussia, A.; Malamou-Mitsi, V.; Panayiotidis, M.I.; et al. Dietary mastic oil extracted from Pistacia lentiscus var. chia suppresses tumor growth in experimental colon cancer models. Sci. Rep. 2017, 7, 3782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spyridopoulou, K. Design and Applications of Bioactive Nanoparticles in Cancer Prevention and Therapy. Ph.D. Thesis, Democritus University of Thrace, Alexandroupolis, Greece, 2018. [Google Scholar]
- Green, D.R.; Ferguson, T.; Zitvogel, L.; Kroemer, G. Immunogenic and tolerogenic cell death. Nat. Rev. Immunol. 2009, 9, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Showalter, A.; Limaye, A.; Oyer, J.L.; Igarashi, R.; Kittipatarin, C.; Copik, A.J.; Khaled, A.R. Cytokines in immunogenic cell death: Applications for cancer immunotherapy. Cytokine 2017, 97, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.P.; Weissman, I.L.; Majeti, R. The CD47-SIRPα pathway in cancer immune evasion and potential therapeutic implications. Curr. Opin. Immunol. 2012, 24, 225–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garg, A.D.; Romano, E.; Rufo, N.; Agostinis, P. Immunogenic versus tolerogenic phagocytosis during anticancer therapy: Mechanisms and clinical translation. Cell Death Differ. 2016, 23, 938–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulgarini, A.; Lampis, S.; Turner, R.J.; Vallini, G. Biomolecular composition of capping layer and stability of biogenic selenium nanoparticles synthesized by five bacterial species. Microb. Biotechnol. 2021, 14, 198–212. [Google Scholar] [CrossRef] [PubMed]
- Kalkavan, H.; Green, D.R. MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 2018, 25, 46–55. [Google Scholar] [CrossRef]
- Youle, R.J.; Strasser, A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef]
- Dadsena, S.; King, L.E.; García-Sáez, A.J. Apoptosis regulation at the mitochondria membrane level. Biochim. Biophys. Acta 2021, 1863, 183716. [Google Scholar] [CrossRef]
- Galluzzi, L.; Vitale, I. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef] [PubMed]
- Asadi, M.; Taghizadeh, S.; Kaviani, E.; Vakili, O.; Taheri-Anganeh, M.; Tahamtan, M.; Savardashtaki, A. Caspase-3; Structure, Function, and Biotechnological Aspects. Biotechnol. Appl. Biochem. 2021, 1, 1–26. [Google Scholar]
- Wang, X.; Chen, M.; Zhou, J.; Zhang, X. HSP27, 70 and 90, anti-apoptotic proteins, in clinical cancer therapy (review). Int. J. Oncol. 2014, 45, 18–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, D.R.; Llambi, F. Cell death signaling. Cold Spring Harb. Perspect. Biol. 2015, 7, a006080. [Google Scholar] [CrossRef]
- Katsogiannou, M.; Andrieu, C.; Rocchi, P. Heat shock protein 27 phosphorylation state is associated with cancer progression. Front. Genet. 2014, 5, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Chaitanya, G.V.; Alexander, J.S.; Babu, P.P. PARP-1 cleavage fragments: Signatures of cell-death proteases in neurodegeneration. Cell Commun. Signal. 2010, 8, 31. [Google Scholar] [CrossRef] [Green Version]
- Yazdi, M.H.; Sepehrizadeh, Z.; Mahdavi, M.; Shahverdi, A.R.; Faramarzi, M.A. Metal, Metalloid, and Oxide Nanoparticles for Therapeutic and Diagnostic Oncology. Nano Biomed. Eng. 2016, 8, 246–267. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Yuan, Q.; Zhu, H.; Li, Y.; Guo, Q.; Wang, Q.; Bi, X.; Gao, X. The suppression of prostate LNCaP cancer cells growth by Selenium nanoparticles through Akt/Mdm2/AR controlled apoptosis. Biomaterials 2011, 32, 6515–6522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, X.; Huang, Z.; Zheng, W.; Fan, C.; Chen, T. Enhancement of cell permeabilization apoptosis-inducing activity of selenium nanoparticles by ATP surface decoration. Nanomed. Nanotechnol. Biol. Med. 2013, 9, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Alkhudhayri, A.A.; Wahab, R.; Siddiqui, M.A.; Ahmad, J. Selenium Nanoparticles Induce Cytotoxicity and Apoptosis in Human Breast Cancer (MCF-7) and Liver (HepG2) Cell Lines. Nanosci. Nanotechnol. Lett. 2020, 12, 324–330. [Google Scholar] [CrossRef]
- Li, H.; Liu, D.; Li, S.; Xue, C. Synthesis and cytotoxicity of selenium nanoparticles stabilized by α-D-glucan from Castanea mollissima Blume. Int. J. Biol. Macromol. 2019, 129, 818–826. [Google Scholar] [CrossRef]
- Yang, F.; Tang, Q.; Zhong, X.; Bai, Y.; Chen, T.; Zhang, Y.; Li, Y.; Zheng, W. Surface decoration by Spirulina polysaccharide enhances the cellular uptake and anticancer efficacy of selenium nanoparticles. Int. J. Nanomed. 2012, 7, 835–844. [Google Scholar]
- Amiri, H.; Hashemy, S.I.; Sabouri, Z.; Javid, H.; Darroudi, M. Green synthesized selenium nanoparticles for ovarian cancer cell apoptosis. Res. Chem. Intermed. 2021, 47, 2539–2556. [Google Scholar] [CrossRef]
- Van Loenhout, J.; Peeters, M.; Bogaerts, A.; Smits, E.; Deben, C. Oxidative stress-inducing anticancer therapies: Taking a closer look at their immunomodulating effects. Antioxidants 2020, 9, 1188. [Google Scholar] [CrossRef] [PubMed]
- Panaretakis, T.; Joza, N.; Modjtahedi, N.; Tesniere, A.; Vitale, I.; Durchschlag, M.; Fimia, G.M.; Kepp, O.; Piacentini, M.; Froehlich, K.-U.; et al. The co-translocation of ERp57 and calreticulin determines the immunogenicity of cell death. Cell Death Differ. 2008, 15, 1499–1509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obeid, M. ERP57 Membrane Translocation Dictates the Immunogenicity of Tumor Cell Death by Controlling the Membrane Translocation of Calreticulin. J. Immunol. 2008, 181, 2533–2543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obeid, M.; Tesniere, A.; Panaretakis, T.; Tufi, R.; Joza, N.; van Endert, P.; Ghiringhelli, F.; Apetoh, L.; Chaput, N.; Flament, C.; et al. Ecto-calreticulin in immunogenic chemotherapy. Immunol. Rev. 2007, 220, 22–34. [Google Scholar] [CrossRef]
- Song, D.; Liu, H.; Wu, J.; Gao, X.; Hao, J.; Fan, D. Insights into the role of ERp57 in cancer. J. Cancer 2021, 12, 2456–2464. [Google Scholar] [CrossRef]
- Zhu, X.; Messer, J.S.; Wang, Y.; Lin, F.; Cham, C.M.; Chang, J.; Billiar, T.R.; Lotze, M.T.; Boone, D.L.; Chang, E.B. Cytosolic HMGB1 controls the cellular autophagy/apoptosis checkpoint during inflammation. J. Clin. Invest. 2015, 125, 1098–1110. [Google Scholar] [CrossRef] [Green Version]
- Martins, I.; Wang, Y.; Michaud, M.; Ma, Y.; Sukkurwala, A.Q.; Shen, S.; Kepp, O.; Métivier, D.; Galluzzi, L.; Perfettini, J.L.; et al. Molecular mechanisms of ATP secretion during immunogenic cell death. Cell Death Differ. 2014, 21, 79–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, S.; Pestka, S.; Jubin, R.G.; Lyu, Y.L.; Tsai, Y.-C.; Liu, L.F. Chemotherapeutics and Radiation Stimulate MHC Class I Expression through Elevated Interferon-beta Signaling in Breast Cancer Cells. PLoS ONE 2012, 7, e32542. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.; Weaver, C.; Weaver, C. Janeway’s Immunobiology, 9th ed.; Garland Science: New York, NY, USA, 2016; ISBN 9781315533247. [Google Scholar]
- Gupta, G.; Borglum, K.; Chen, H. Immunogenic Cell Death: A Step Ahead of Autophagy in Cancer Therapy. J. Cancer Immunol. 2021, 3, 47–59. [Google Scholar]
- Plotkin, S.; Orenstein, W.; Offit, P.; Edwards, K.M. Plotkin’s Vaccines; Elsevier: Amsterdam, The Netherlands, 2018; ISBN 9780323357616. [Google Scholar]
- Hollingsworth, R.E.; Jansen, K. Turning the corner on therapeutic cancer vaccines. NPJ Vaccines 2019, 4, 1–10. [Google Scholar] [CrossRef]
- Qi, J.; Jin, F.; Xu, X.; Du, Y. Combination Cancer Immunotherapy of Nanoparticle-Based Immunogenic Cell Death Inducers and Immune Checkpoint Inhibitors. Int. J. Nanomed. 2021, 16, 1435–1456. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Sun, I.-C.; Hwang, H.S.; Shim, M.K.; Yoon, H.Y.; Kim, K. Rediscovery of nanoparticle-based therapeutics: Boosting immunogenic cell death for potential application in cancer immunotherapy. J. Mater. Chem. B 2021, 9, 3983–4001. [Google Scholar] [CrossRef] [PubMed]
- Aindelis, G.; Tiptiri-Kourpeti, A.; Lampri, E.; Spyridopoulou, K.; Lamprianidou, E.; Kotsianidis, I.; Ypsilantis, P.; Pappa, A.; Chlichlia, K. Immune responses raised in an experimental colon carcinoma model following oral administration of Lactobacillus casei. Cancers 2020, 12, 368. [Google Scholar] [CrossRef] [PubMed] [Green Version]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spyridopoulou, K.; Aindelis, G.; Pappa, A.; Chlichlia, K. Anticancer Activity of Biogenic Selenium Nanoparticles: Apoptotic and Immunogenic Cell Death Markers in Colon Cancer Cells. Cancers 2021, 13, 5335. https://doi.org/10.3390/cancers13215335
Spyridopoulou K, Aindelis G, Pappa A, Chlichlia K. Anticancer Activity of Biogenic Selenium Nanoparticles: Apoptotic and Immunogenic Cell Death Markers in Colon Cancer Cells. Cancers. 2021; 13(21):5335. https://doi.org/10.3390/cancers13215335
Chicago/Turabian StyleSpyridopoulou, Katerina, Georgios Aindelis, Aglaia Pappa, and Katerina Chlichlia. 2021. "Anticancer Activity of Biogenic Selenium Nanoparticles: Apoptotic and Immunogenic Cell Death Markers in Colon Cancer Cells" Cancers 13, no. 21: 5335. https://doi.org/10.3390/cancers13215335
APA StyleSpyridopoulou, K., Aindelis, G., Pappa, A., & Chlichlia, K. (2021). Anticancer Activity of Biogenic Selenium Nanoparticles: Apoptotic and Immunogenic Cell Death Markers in Colon Cancer Cells. Cancers, 13(21), 5335. https://doi.org/10.3390/cancers13215335