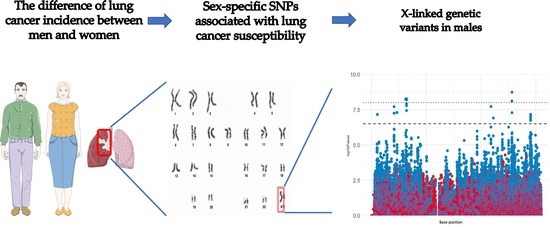
Identification of Genetic Variants Associated with Sex-Specific Lung-Cancer Risk
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Accession and Categorization
2.2. Association Analysis
2.3. Bioinformatics Analysis for DMD Expression and Mutation
2.4. GO/KEGG Pathway Enrichment Analysis
2.5. Statistical Analysis
3. Results
3.1. Identification of Sex-Specific SNPs Associated with Lung Cancer Susceptibility
3.2. Interactions between X Chromosome SNPs in Male Lung Cancer Risk
3.3. Effect of Sex-Specific Lung Cancer Risk SNPs on DMD Expression
3.4. DMD Could Affect CD4+ T Cell Infiltration in LUSC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bain, C.; Feskanich, D.; Speizer, F.E.; Thun, M.; Hertzmark, E.; Rosner, B.A.; Colditz, G.A. Lung cancer rates in men and women with comparable histories of smoking. J. Natl. Cancer Inst. 2004, 96, 826–834. [Google Scholar] [CrossRef] [Green Version]
- Zang, E.A.; Wynder, E.L. Differences in lung cancer risk between men and women: Examination of the evidence. J. Natl. Cancer Inst. 1996, 88, 183–192. [Google Scholar] [CrossRef]
- Bosse, Y.; Amos, C.I. A Decade of GWAS Results in Lung Cancer. Cancer Epidemiol. Biomark. Prev. 2018, 27, 363–379. [Google Scholar] [CrossRef] [Green Version]
- Li, C.H.; Prokopec, S.D.; Sun, R.X.; Yousif, F.; Schmitz, N.; Subtypes, P.T.; Clinical, T.; Boutros, P.C.; Consortium, P. Sex differences in oncogenic mutational processes. Nat. Commun. 2020, 11, 4330. [Google Scholar] [CrossRef]
- Dunford, A.; Weinstock, D.M.; Savova, V.; Schumacher, S.E.; Cleary, J.P.; Yoda, A.; Sullivan, T.J.; Hess, J.M.; Gimelbrant, A.A.; Beroukhim, R.; et al. Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet. 2017, 49, 10–16. [Google Scholar] [CrossRef] [Green Version]
- Haupt, S.; Caramia, F.; Herschtal, A.; Soussi, T.; Lozano, G.; Chen, H.; Liang, H.; Speed, T.P.; Haupt, Y. Identification of cancer sex-disparity in the functional integrity of p53 and its X chromosome network. Nat. Commun. 2019, 10, 5385. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Kinnersley, B.; Wrensch, M.R.; Eckel-Passow, J.E.; Armstrong, G.; Rice, T.; Chen, Y.; Wiencke, J.K.; McCoy, L.S.; Hansen, H.M.; et al. Sex-specific glioma genome-wide association study identifies new risk locus at 3p21.31 in females, and finds sex-differences in risk at 8q24.21. Sci. Rep. 2018, 8, 7352. [Google Scholar] [CrossRef] [Green Version]
- Zuo, X.Y.; Feng, Q.S.; Sun, J.; Wei, P.P.; Chin, Y.M.; Guo, Y.M.; Xia, Y.F.; Li, B.; Xia, X.J.; Jia, W.H.; et al. X-chromosome association study reveals genetic susceptibility loci of nasopharyngeal carcinoma. Biol. Sex Differ. 2019, 10, 13. [Google Scholar] [CrossRef]
- Chun, K.A.; Kocarnik, J.M.; Hardikar, S.S.; Robinson, J.R.; Berndt, S.I.; Chan, A.T.; Figueiredo, J.C.; Lindor, N.M.; Song, M.; Schoen, R.E.; et al. Leptin gene variants and colorectal cancer risk: Sex-specific associations. PLoS ONE 2018, 13, e0206519. [Google Scholar] [CrossRef]
- Roshyara, N.R.; Scholz, M. fcGENE: A versatile tool for processing and transforming SNP datasets. PLoS ONE 2014, 9, e97589. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Landi, M.T.; Consonni, D.; Rotunno, M.; Bergen, A.W.; Goldstein, A.M.; Lubin, J.H.; Goldin, L.; Alavanja, M.; Morgan, G.; Subar, A.F.; et al. Environment and Genetics in Lung cancer Etiology (EAGLE) study: An integrative population-based case-control study of lung cancer. BMC Public Health 2008, 8, 203. [Google Scholar] [CrossRef] [Green Version]
- Prorok, P.C.; Andriole, G.L.; Bresalier, R.S.; Buys, S.S.; Chia, D.; Crawford, E.D.; Fogel, R.; Gelmann, E.P.; Gilbert, F.; Hasson, M.A.; et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control. Clin. Trials 2000, 21, 273S–309S. [Google Scholar] [CrossRef]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhsng, C.Z.; Wala, J.; Mermel, C.H.; et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013, 45, 1134–1140. [Google Scholar] [CrossRef] [Green Version]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011, 12, R41. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stapelfeld, C.; Dammann, C.; Maser, E. Sex-specificity in lung cancer risk. Int. J. Cancer 2020, 146, 2376–2382. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Park, D.Y.; Kim, M.Y.; Kang, C. Synergistic triad epistasis of epigenetic H3K27me modifier genes, EZH2, KDM6A, and KDM6B, in gastric cancer susceptibility. Gastric Cancer 2019, 22, 640–644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.J.; Tseng, Y.K.; Lo, Y.H.; Wu, P.C.; Lee, J.H.; Liou, H.H.; Liang, C.C.; Yang, C.M.; Wang, C.C.; Yen, L.M.; et al. Association of SDF-1 and CXCR4 Polymorphisms with Susceptibility to Oral and Pharyngeal Squamous Cell Carcinoma. Anticancer Res. 2019, 39, 2891–2902. [Google Scholar] [CrossRef]
- Jones, L.; Naidoo, M.; Machado, L.R.; Anthony, K. The Duchenne muscular dystrophy gene and cancer. Cell Oncol. (Dordr.) 2021, 44, 19–32. [Google Scholar] [CrossRef]




| SNP | BP | A1 | F_A | F_U | A2 | CHISQ | P * | OR | Gene |
|---|---|---|---|---|---|---|---|---|---|
| rs6529797 | 5454436 | T | 0.1709 | 0.08176 | G | 29.07 | 7 × 10−8 | 2.315 | - |
| rs4364769 | 5462201 | T | 0.1709 | 0.08176 | G | 29.07 | 7 × 10−8 | 2.315 | - |
| rs17313971 | 20597131 | G | 0.1262 | 0.04822 | T | 31.55 | 2 × 10−8 | 2.851 | - |
| rs59299065 | 20600981 | G | 0.1182 | 0.04507 | C | 29.49 | 5.6 × 10−8 | 2.84 | - |
| rs7879756 | 20601076 | A | 0.1182 | 0.04507 | C | 29.49 | 5.6 × 10−8 | 2.84 | - |
| rs35550069 | 20602845 | C | 0.1182 | 0.04507 | T | 29.49 | 5.6 × 10−8 | 2.84 | - |
| rs5990896 | 20603703 | G | 0.1182 | 0.04507 | A | 29.49 | 5.6 × 10−8 | 2.84 | - |
| rs7883926 | 23285507 | C | 0 | 0.04612 | A | 29.7 | 5.1 × 10−8 | 0 | PTCHD1-AS |
| rs6526263 | 23286674 | G | 0 | 0.04612 | T | 29.7 | 5.1 × 10−8 | 0 | PTCHD1-AS |
| rs55803048 | 31273345 | A | 0.1821 | 0.08386 | G | 33.88 | 5.9 × 10−9 | 2.433 | DMD |
| rs7057271 | 31277489 | T | 0.1837 | 0.08491 | A | 33.96 | 5.6 × 10−9 | 2.426 | DMD |
| rs5971605 | 31977216 | C | 0.3035 | 0.1782 | T | 33.74 | 6.3 × 10−9 | 2.01 | DMD |
| rs5972468 | 31978713 | T | 0.3035 | 0.1782 | C | 33.74 | 6.3 × 10−9 | 2.01 | DMD |
| rs5972469 | 31978946 | C | 0.3035 | 0.1782 | T | 33.74 | 6.3 × 10−9 | 2.01 | DMD |
| rs60546832 | 31983160 | T | 0.3035 | 0.1803 | C | 32.49 | 1.2 × 10−8 | 1.981 | DMD |
| rs62587743 | 31985166 | T | 0.3514 | 0.218 | C | 34.06 | 5.3 × 10−9 | 1.943 | DMD |
| rs7051329 | 31985724 | T | 0.2971 | 0.1761 | G | 31.84 | 1.7 × 10−8 | 1.978 | DMD |
| rs67632566 | 31988691 | T | 0.2939 | 0.1761 | C | 30.31 | 3.7 × 10−8 | 1.948 | DMD |
| rs57391430 | 109757735 | G | 0.2812 | 0.4203 | A | 31.59 | 1.9 × 10−8 | 0.5394 | - |
| rs145211462 | 129041862 | T | 0.03834 | 0.1279 | C | 36.14 | 1.8 × 10−9 | 0.2719 | AL008633.1 |
| rs200018042 | 129048102 | G | 0.1565 | 0.282 | C | 33.34 | 7.7 × 10−9 | 0.4726 | AL008633.1 |
| rs6637526 | 129048867 | G | 0.2604 | 0.3931 | A | 29.64 | 5.2 × 10−8 | 0.5436 | AL008633.1 |
| rs182684417 | 146125695 | G | 0.1757 | 0.08595 | A | 28.53 | 9.2 × 10−8 | 2.267 | - |
| rs62601607 | 146134454 | T | 0.1534 | 0.06918 | C | 29.1 | 6.9 × 10−8 | 2.437 | - |
| Peak SNPs | Genotype | Cases (n = 313) | Controls (n = 477) | OR * | P |
|---|---|---|---|---|---|
| rs6529797 | T | 54 | 39 | 1.56 | <0.01 |
| G | 259 | 438 | |||
| rs17313971 | G | 44 | 27 | 1.65 | <0.01 |
| T | 269 | 450 | |||
| rs7883926 | A | 313 | 455 | NA | <0.01 |
| C | 0 | 22 | |||
| rs55803048 | A | 57 | 40 | 1.59 | <0.01 |
| G | 256 | 437 | |||
| rs145211462 | C | 301 | 416 | 2.55 | <0.01 |
| T | 12 | 61 | |||
| rs62601607 | T | 48 | 33 | 1.59 | <0.01 |
| C | 265 | 444 |
| SNP | Genotype | Cases | Controls | OR * | P |
|---|---|---|---|---|---|
| rs6529797 | T | 51 | 71 | 1.14 | 0.31 |
| G | 164 | 282 | |||
| rs17313971 | G | 25 | 39 | 1.04 | 0.83 |
| T | 190 | 314 | |||
| rs7883926 | A | 202 | 321 | 1.34 | 0.19 |
| C | 13 | 32 | |||
| rs55803048 | A | 47 | 69 | 1.09 | 0.51 |
| G | 168 | 284 | |||
| rs145211462 | C | 189 | 304 | 1.11 | 0.54 |
| T | 26 | 49 | |||
| rs62601607 | T | 47 | 74 | 1.03 | 0.8 |
| C | 168 | 279 |
| SNP * | Age | Resistance Allele | Risk Allele | P |
|---|---|---|---|---|
| rs6529797 | Under 65 | 134 | 19 | 0.04 |
| Over 65 | 125 | 35 | ||
| rs17313971 | Under 65 | 129 | 24 | 0.52 |
| Over 65 | 140 | 20 | ||
| rs55803048 | Under 65 | 129 | 24 | 0.31 |
| Over 65 | 127 | 33 | ||
| rs145211462 | Under 65 | 10 | 143 | 0.02 |
| Over 65 | 2 | 158 | ||
| rs62601607 | Under 65 | 125 | 28 | 0.16 |
| Over 65 | 140 | 20 |
| Combination * | Genotype | 2 by 2 Risk Alleles Combination | |||||
|---|---|---|---|---|---|---|---|
| Male | Female | ||||||
| Cancer | Control | Odds | Cancer | Control | Odds | ||
| X1_2 | G_T | 220 | 411 | 0.53 | 190 | 314 | 0.61 |
| X1_3 | T_A | 54 | 39 | 1.38 | None | ||
| X1_4 | T_A | 12 | 2 | 6 | None | ||
| X1_5 | T_C | 52 | 31 | 1.68 | None | ||
| X1_6 | T_T | 5 | 2 | 2.5 | None | ||
| X2_3 | G_A | 44 | 26 | 1.70 | None | ||
| X2_4 | G_A | 10 | 2 | 5 | None | ||
| X2_5 | G_C | 42 | 26 | 1.62 | None | ||
| X2_6 | G_T | 6 | 2 | 3 | None | ||
| X3_4 | A_A | 57 | 39 | 1.46 | 47 | 69 | 0.68 |
| X3_5 | A_C | 301 | 397 | 0.76 | 189 | 304 | 0.62 |
| X3_6 | A_T | 48 | 31 | 1.55 | 47 | 74 | 0.64 |
| X4_5 | A_C | 57 | 33 | 1.73 | None | ||
| X4_6 | A_T | 8 | 4 | 2 | None | ||
| X5_6 | C_T | 47 | 28 | 1.68 | None | ||
| Combination * | Genotype | Cancer | Control | Odds |
|---|---|---|---|---|
| X1_2_3 | T_G_A | 5 | 0 | NA |
| X1_2_4 | T_G_A | 1 | 0 | NA |
| X1_2_5 | T_G_C | 5 | 0 | NA |
| X1_2_6 | - | - | - | - |
| X1_3_4 | T_A_A | 12 | 2 | 6 |
| X1_3_5 | T_A_C | 52 | 31 | 1.68 |
| X1_3_6 | T_A_T | 52 | 31 | 2.5 |
| X1_4_5 | T_A_C | 12 | 2 | 6 |
| X1_4_6 | - | - | - | - |
| X1_5_6 | T_C_T | 5 | 2 | 2.5 |
| X2_3_4 | G_A_A | 5 | 2 | 5 |
| X2_3_5 | G_A_C | 42 | 25 | 1.68 |
| X2_3_6 | G_A_T | 6 | 2 | 3 |
| X3_4_5 | A_A_C | 57 | 32 | 1.78 |
| X3_4_6 | - | - | - | - |
| X4_5_6 | A_C_T | 8 | 3 | 2.67 |
| Dataset | Lung Cancer Subtype | Endpoint | Probe ID | N | COX p-Value | HR,95% CI [Lower-Upper Bound] * |
|---|---|---|---|---|---|---|
| GSE31210 | Adenocarcinoma | Relapse-free survival | 203881_s_at | 204 | <0.01 | 0.46 [0.32–0.67] |
| GSE13213 | Adenocarcinoma | Overall survival | A_24_P185854 | 117 | <0.01 | 0.64 [0.50–0.82] |
| GSE31210 | Adenocarcinoma | Overall survival | 203881_s_at | 204 | <0.01 | 0.46 [0.29–0.75] |
| GSE31210 | Adenocarcinoma | Relapse-free survival | 207660_at | 204 | <0.01 | 0.53 [0.34–0.82] |
| jacob-00182-UM | Adenocarcinoma | Overall survival | 203881_s_at | 178 | 0.01 | 0.76 [0.61–0.94] |
| GSE8894 | # NSCLC | Relapse-free survival | 234752_x_at | 138 | 0.04 | 0.17 [0.03–0.91] |
| GSE31210 | Adenocarcinoma | Relapse-free survival | 208086_s_at | 204 | 0.06 | 0.62 [0.37–1.01] |
| GSE8894 | NSCLC | Relapse-free survival | 207660_at | 138 | 0.08 | 0.00 [0.00–3.61] |
| GSE3141 | NSCLC | Overall survival | 203881_s_at | 111 | 0.11 | 0.78 [0.58–1.06] |
| jacob-00182-MSK | Adenocarcinoma | Overall survival | 203881_s_at | 104 | 0.16 | 0.78 [0.55–1.10] |
| jacob-00182-CANDF | Adenocarcinoma | Overall survival | 203881_s_at | 82 | 0.16 | 0.75 [0.50–1.13] |
| GSE13213 | Adenocarcinoma | Overall survival | A_24_P34186 | 117 | 0.17 | 0.76 [0.51–1.13] |
| GSE31210 | Adenocarcinoma | Overall survival | 208086_s_at | 204 | 0.18 | 0.64 [0.33–1.23] |
| GSE3141 | NSCLC | Overall survival | 207660_at | 111 | 0.20 | 0.76 [0.50–1.15] |
| jacob-00182-MSK | Adenocarcinoma | Overall survival | 208086_s_at | 104 | 0.22 | 0.82 [0.60–1.12] |
| GSE31210 | Adenocarcinoma | Overall survival | 207660_at | 204 | 0.26 | 0.69 [0.36–1.32] |
| MICHIGAN-LC | Adenocarcinoma | Overall survival | M18533_at | 86 | 0.28 | 0.76 [0.46–1.25] |
| jacob-00182-UM | Adenocarcinoma | Overall survival | 207660_at | 178 | 0.28 | 0.69 [0.35–1.35] |
| GSE17710 | Squamous cell carcinoma | Relapse-free survival | 26567 | 56 | 0.28 | 1.70 [0.65–4.47] |
| GSE17710 | Squamous cell carcinoma | Overall survival | 26567 | 56 | 0.31 | 1.69 [0.62–4.64] |
| GSE17710 | Squamous cell carcinoma | Relapse-free survival | 3354 | 56 | 0.32 | 1.53 [0.67–3.50] |
| GSE14814 | NSCLC | Disease-specific survival | 208086_s_at | 90 | 0.35 | 2.10 [0.44–9.94] |
| jacob-00182-MSK | Adenocarcinoma | Overall survival | 207660_at | 104 | 0.36 | 0.62 [0.22–1.75] |
| GSE17710 | Squamous cell carcinoma | Relapse-free survival | 11043 | 56 | 0.37 | 1.15 [0.85–1.56] |
| jacob-00182-HLM | Adenocarcinoma | Overall survival | 207660_at | 79 | 0.38 | 0.64 [0.24–1.73] |
| GSE4573 | Squamous cell carcinoma | Overall survival | 208086_s_at | 129 | 0.40 | 0.83 [0.55–1.28] |
| GSE17710 | Squamous cell carcinoma | Overall survival | 3354 | 56 | 0.45 | 1.39 [0.58–3.34] |
| GSE31210 | Adenocarcinoma | Overall survival | 234752_x_at | 204 | 0.46 | 0.88 [0.63–1.24] |
| GSE3141 | NSCLC | Overall survival | 234752_x_at | 111 | 0.47 | 1.07 [0.89–1.29] |
| MICHIGAN-LC | Adenocarcinoma | Overall survival | S81419_at | 86 | 0.50 | 0.73 [0.29–1.84] |
| GSE17710 | Squamous cell carcinoma | Overall survival | 8673 | 56 | 0.53 | 0.80 [0.39–1.63] |
| jacob-00182-CANDF | Adenocarcinoma | Overall survival | 207660_at | 82 | 0.53 | 0.77 [0.35–1.74] |
| jacob-00182-HLM | Adenocarcinoma | Overall survival | 203881_s_at | 79 | 0.54 | 0.92 [0.69–1.21] |
| GSE4716-GPL3696 | NSCLC | Overall survival | 3336 | 50 | 0.54 | 0.80 [0.40–1.62] |
| GSE3141 | NSCLC | Overall survival | 208086_s_at | 111 | 0.60 | 0.89 [0.58–1.37] |
| GSE4573 | Squamous cell carcinoma | Overall survival | 207660_at | 129 | 0.61 | 1.17 [0.63–2.16] |
| GSE14814 | NSCLC | Overall survival | 208086_s_at | 90 | 0.63 | 1.48 [0.30–7.36] |
| GSE17710 | Squamous cell carcinoma | Relapse-free survival | 8673 | 56 | 0.64 | 0.85 [0.41–1.73] |
| GSE17710 | Squamous cell carcinoma | Overall survival | 11043 | 56 | 0.67 | 1.06 [0.80–1.41] |
| GSE31210 | Adenocarcinoma | Relapse-free survival | 234752_x_at | 204 | 0.68 | 0.94 [0.71–1.25] |
| GSE4573 | Squamous cell carcinoma | Overall survival | 203881_s_at | 129 | 0.72 | 0.95 [0.72–1.25] |
| GSE8894 | NSCLC | Relapse-free survival | 208086_s_at | 138 | 0.74 | 0.87 [0.38–1.99] |
| GSE14814 | NSCLC | Disease-specific survival | 203881_s_at | 90 | 0.77 | 1.10 [0.58–2.07] |
| GSE14814 | NSCLC | Overall survival | 207660_at | 90 | 0.82 | 0.88 [0.28–2.78] |
| GSE8894 | NSCLC | Relapse-free survival | 203881_s_at | 138 | 0.87 | 0.99 [0.83–1.17] |
| GSE14814 | NSCLC | Disease-specific survival | 207660_at | 90 | 0.93 | 1.06 [0.29–3.90] |
| jacob-00182-UM | Adenocarcinoma | Overall survival | 208086_s_at | 178 | 0.96 | 1.00 [0.83–1.20] |
| GSE14814 | NSCLC | Overall survival | 203881_s_at | 90 | 0.96 | 1.01 [0.57–1.79] |
| jacob-00182-HLM | Adenocarcinoma | Overall survival | 208086_s_at | 79 | 0.99 | 1.00 [0.78–1.29] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, X.; Young, S.; Morahan, G. Identification of Genetic Variants Associated with Sex-Specific Lung-Cancer Risk. Cancers 2021, 13, 6379. https://doi.org/10.3390/cancers13246379
Shi X, Young S, Morahan G. Identification of Genetic Variants Associated with Sex-Specific Lung-Cancer Risk. Cancers. 2021; 13(24):6379. https://doi.org/10.3390/cancers13246379
Chicago/Turabian StyleShi, Xiaoshun, Sylvia Young, and Grant Morahan. 2021. "Identification of Genetic Variants Associated with Sex-Specific Lung-Cancer Risk" Cancers 13, no. 24: 6379. https://doi.org/10.3390/cancers13246379
APA StyleShi, X., Young, S., & Morahan, G. (2021). Identification of Genetic Variants Associated with Sex-Specific Lung-Cancer Risk. Cancers, 13(24), 6379. https://doi.org/10.3390/cancers13246379