Circulating Cell-Free DNA Combined to Magnetic Resonance Imaging for Early Detection of HCC in Patients with Liver Cirrhosis
Abstract
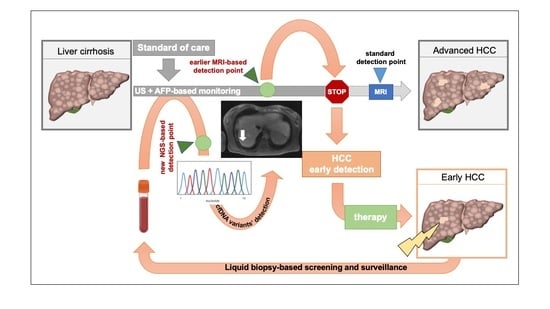
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Clinicopathological Characteristics of the Patients
2.2. Quantification of cfDNA in Cirrhotic Patients and Healthy Donors
2.3. Somatic Mutation Analysis
2.4. Imaging Features in Patients Carrying cfDNA Variants
2.5. Follow-Up Study
2.6. Association of cfDNA Levels and Variants with Patient’s Clinicopathological Characteristics
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Image Acquisition
4.3. Image Analysis
4.4. DNA Isolation, Library Preparation and Next-Generation Sequencing (NGS)
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Xing, F.; Yeo, Y.H.; Jin, M.; Le, R.; Le, M.; Jin, M.; Henry, L.; Cheung, R.; Nguyen, M.H. Only one-third of hepatocellular carcinoma cases are diagnosed via screening or surveillance: A systematic review and meta-analysis. Eur. J. Gastroenterol. Hepatol. 2020, 32, 406–419. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Burroughs, A.; Bruix, J. Hepatocellular carcinoma. Lancet 2003, 362, 1907–1917. [Google Scholar] [CrossRef] [Green Version]
- Forner, A.; Llovet, J.M.; Bruix, J. Hepatocellular carcinoma. Lancet 2012, 379, 1245–1255. [Google Scholar] [CrossRef]
- Ramalho, M.; Matos, A.P.; AlObaidy, M.; Velloni, F.; Altun, E.; Semelka, R.C. Magnetic resonance imaging of the cirrhotic liver: Diagnosis of hepatocellular carcinoma and evaluation of response to treatment—Part 2. Radiol. Bras. 2017, 50, 115–125. [Google Scholar] [CrossRef] [Green Version]
- Yu, N.C.; Chaudhari, V.; Raman, S.S.; Lassman, C.; Tong, M.J.; Busuttil, R.W.; Lu, D.S. CT and MRI improve detection of hepatocellular carcinoma, compared with ultrasound alone, in patients with cirrhosis. Clin. Gastroenterol. Hepatol. 2011, 9, 161–167. [Google Scholar] [CrossRef]
- Alunni-Fabbroni, M.; Muller, V.; Fehm, T.; Janni, W.; Rack, B. Monitoring in metastatic breast cancer: Is imaging outdated in the era of circulating tumor cells? Breast Care 2014, 9, 16–21. [Google Scholar] [CrossRef] [Green Version]
- Alix-Panabieres, C.; Schwarzenbach, H.; Pantel, K. Circulating tumor cells and circulating tumor DNA. Annu. Rev. Med. 2012, 63, 199–215. [Google Scholar] [CrossRef]
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochim. Biophys. Acta 2007, 1775, 181–232. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, N.; Sakaida, I.; Moribe, T.; Fujita, N.; Miura, T.; Stark, M.; Tamatsukuri, S.; Ishitsuka, H.; Uchida, K.; Terai, S.; et al. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis C virus-associated hepatocellular carcinoma. Anticancer Res. 2006, 26, 4713–4719. [Google Scholar] [PubMed]
- Tokuhisa, Y.; Iizuka, N.; Sakaida, I.; Moribe, T.; Fujita, N.; Miura, T.; Tamatsukuri, S.; Ishitsuka, H.; Uchida, K.; Terai, S.; et al. Circulating cell-free DNA as a predictive marker for distant metastasis of hepatitis C virus-related hepatocellular carcinoma. Br. J. Cancer 2007, 97, 1399–1403. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Patel, K.M.; Tsui, D.W. The translational potential of circulating tumour DNA in oncology. Clin. Biochem. 2015, 48, 957–961. [Google Scholar] [CrossRef]
- Xu, R.H.; Wei, W.; Krawczyk, M.; Wang, W.; Luo, H.; Flagg, K.; Yi, S.; Shi, W.; Quan, Q.; Li, K.; et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017, 16, 1155–1161. [Google Scholar] [CrossRef]
- Ye, Q.; Ling, S.; Zheng, S.; Xu, X. Liquid biopsy in hepatocellular carcinoma: Circulating tumor cells and circulating tumor DNA. Mol. Cancer 2019, 18, 114. [Google Scholar] [CrossRef]
- Yang, J.D.; Liu, M.C.; Kisiel, J.B. Circulating Tumor DNA and Hepatocellular Carcinoma. In Seminars in Liver Disease; Thieme Medical Publishers: New York, NY, USA, 2019; Volume 39, pp. 452–462. [Google Scholar]
- Piciocchi, M.; Cardin, R.; Vitale, A.; Vanin, V.; Giacomin, A.; Pozzan, C.; Maddalo, G.; Cillo, U.; Guido, M.; Farinati, F. Circulating free DNA in the progression of liver damage to hepatocellular carcinoma. Hepatol. Int. 2013, 7, 1050–1057. [Google Scholar] [CrossRef]
- Ren, N.; Ye, Q.H.; Qin, L.X.; Zhang, B.H.; Liu, Y.K.; Tang, Z.Y. Circulating DNA level is negatively associated with the long-term survival of hepatocellular carcinoma patients. World J. Gastroenterol. 2006, 12, 3911–3914. [Google Scholar] [CrossRef]
- Huang, Z.; Hua, D.; Hu, Y.; Cheng, Z.; Zhou, X.; Xie, Q.; Wang, Q.; Wang, F.; Du, X.; Zeng, Y. Quantitation of plasma circulating DNA using quantitative PCR for the detection of hepatocellular carcinoma. Pathol. Oncol. Res. 2012, 18, 271–276. [Google Scholar]
- Kaseb, A.O.; Sanchez, N.S.; Sen, S.; Kelley, R.K.; Tan, B.; Bocobo, A.G.; Lim, K.H.; Abdel-Wahab, R.; Uemura, M.; Pestana, R.C.; et al. Molecular Profiling of Hepatocellular Carcinoma Using Circulating Cell-Free DNA. Clin. Cancer Res. 2019, 25, 6107–6118. [Google Scholar] [CrossRef] [Green Version]
- Alunni-Fabbroni, M.; Ronsch, K.; Huber, T.; Cyran, C.C.; Seidensticker, M.; Mayerle, J.; Pech, M.; Basu, B.; Verslype, C.; Benckert, J.; et al. Circulating DNA as prognostic biomarker in patients with advanced hepatocellular carcinoma: A translational exploratory study from the SORAMIC trial. J. Transl. Med. 2019, 17, 328. [Google Scholar] [CrossRef] [Green Version]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Bolondi, L.; Gaiani, S.; Celli, N.; Golfieri, R.; Grigioni, W.F.; Leoni, S.; Venturi, A.M.; Piscaglia, F. Characterization of small nodules in cirrhosis by assessment of vascularity: The problem of hypovascular hepatocellular carcinoma. Hepatology 2005, 42, 27–34. [Google Scholar] [CrossRef]
- Ioannou, G.N.; Splan, M.F.; Weiss, N.S.; McDonald, G.B.; Beretta, L.; Lee, S.P. Incidence and predictors of hepatocellular carcinoma in patients with cirrhosis. Clin. Gastroenterol. Hepatol. 2007, 5, 938–945.e4. [Google Scholar] [CrossRef]
- Gastrointestinal, I.; Bashir, M.R.; Horowitz, J.M.; Kamel, I.R.; Arif-Tiwari, H.; Asrani, S.K.; Chernyak, V.; Goldstein, A.; Grajo, J.R.; Hindman, N.M.; et al. ACR Appropriateness Criteria(R) Chronic Liver Disease. J. Am. Coll. Radiol. 2020, 17, S70–S80. [Google Scholar]
- Zhang, B.H.; Yang, B.H.; Tang, Z.Y. Randomized controlled trial of screening for hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2004, 130, 417–422. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. Electronic address eee, European Association for the Study of the L. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [Green Version]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar]
- Bolondi, L. Screening for hepatocellular carcinoma in cirrhosis. J. Hepatol. 2003, 39, 1076–1084. [Google Scholar]
- Singal, A.; Volk, M.L.; Waljee, A.; Salgia, R.; Higgins, P.; Rogers, M.A.; Marrero, J.A. Meta-analysis: Surveillance with ultrasound for early-stage hepatocellular carcinoma in patients with cirrhosis. Aliment. Pharmacol. Ther. 2009, 30, 37–47. [Google Scholar]
- Renzulli, M.; Biselli, M.; Brocchi, S.; Granito, A.; Vasuri, F.; Tovoli, F.; Sessagesimi, E.; Piscaglia, F.; D’Errico, A.; Bolondi, L.; et al. New hallmark of hepatocellular carcinoma, early hepatocellular carcinoma and high-grade dysplastic nodules on Gd-EOB-DTPA MRI in patients with cirrhosis: A new diagnostic algorithm. Gut 2018, 67, 1674–1682. [Google Scholar] [CrossRef]
- Karlas, T.; Weise, L.; Kuhn, S.; Krenzien, F.; Mehdorn, M.; Petroff, D.; Linder, N.; Schaudinn, A.; Busse, H.; Keim, V.; et al. Correlation of cell-free DNA plasma concentration with severity of non-alcoholic fatty liver disease. J. Transl. Med. 2017, 15, 106. [Google Scholar] [CrossRef]
- Xia, W.Y.; Gao, L.; Dai, E.H.; Chen, D.; Xie, E.F.; Yang, L.; Zhang, S.C.; Zhang, B.F.; Xu, J.; Pan, S.Y. Liquid biopsy for non-invasive assessment of liver injury in hepatitis B patients. World J. Gastroenterol. 2019, 25, 3985–3995. [Google Scholar]
- Campbell, K.J.; Tait, S.W.G. Targeting BCL-2 regulated apoptosis in cancer. Open Biol. 2018, 8, 180002. [Google Scholar]
- Rampino, N.; Yamamoto, H.; Ionov, Y.; Li, Y.; Sawai, H.; Reed, J.C.; Perucho, M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science 1997, 275, 967–969. [Google Scholar] [CrossRef]
- Nault, J.C.; Bioulac-Sage, P.; Zucman-Rossi, J. Hepatocellular benign tumors-from molecular classification to personalized clinical care. Gastroenterology 2013, 144, 888–902. [Google Scholar] [CrossRef]
- Bluteau, O.; Jeannot, E.; Bioulac-Sage, P.; Marques, J.M.; Blanc, J.F.; Bui, H.; Beaudoin, J.C.; Franco, D.; Balabaud, C.; Laurent-Puig, P.; et al. Bi-allelic inactivation of TCF1 in hepatic adenomas. Nat. Genet. 2002, 32, 312–315. [Google Scholar]
- Cereghini, S.; Yaniv, M.; Cortese, R. Hepatocyte dedifferentiation and extinction is accompanied by a block in the synthesis of mRNA coding for the transcription factor HNF1/LFB1. EMBO J. 1990, 9, 2257–2263. [Google Scholar]
- Zeng, X.; Lin, Y.; Yin, C.; Zhang, X.; Ning, B.F.; Zhang, Q.; Zhang, J.P.; Qiu, L.; Qin, X.R.; Chen, Y.X.; et al. Recombinant adenovirus carrying the hepatocyte nuclear factor-1alpha gene inhibits hepatocellular carcinoma xenograft growth in mice. Hepatology 2011, 54, 2036–2047. [Google Scholar] [CrossRef]
- Luo, Z.; Li, Y.; Wang, H.; Fleming, J.; Li, M.; Kang, Y.; Zhang, R.; Li, D. Hepatocyte nuclear factor 1A (HNF1A) as a possible tumor suppressor in pancreatic cancer. PLoS ONE 2015, 10, e0121082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephens, P.J.; Tarpey, P.S.; Davies, H.; Van Loo, P.; Greenman, C.; Wedge, D.C.; Nik-Zainal, S.; Martin, S.; Varela, I.; Bignell, G.R.; et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012, 486, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Boultwood, J.; Perry, J.; Pellagatti, A.; Fernandez-Mercado, M.; Fernandez-Santamaria, C.; Calasanz, M.J.; Larrayoz, M.J.; Garcia-Delgado, M.; Giagounidis, A.; Malcovati, L.; et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia 2010, 24, 1062–1065. [Google Scholar] [CrossRef]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, M. Functional and cancer genomics of ASXL family members. Br. J. Cancer 2013, 109, 299–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paweletz, C.P.; Sacher, A.G.; Raymond, C.K.; Alden, R.S.; O’Connell, A.; Mach, S.L.; Kuang, Y.; Gandhi, L.; Kirschmeier, P.; English, J.M.; et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin. Cancer Res. 2016, 22, 915–922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banini, B.A.; Sanyal, A.J. The use of cell free DNA in the diagnosis of HCC. Hepatoma Res. 2019, 5. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Takeda, H.; Takai, A.; Matsumoto, T.; Kakiuchi, N.; Yokoyama, A.; Yoshida, K.; Kaido, T.; Uemoto, S.; Minamiguchi, S.; et al. Comprehensive analysis of genetic aberrations linked to tumorigenesis in regenerative nodules of liver cirrhosis. J. Gastroenterol. 2019, 54, 628–640. [Google Scholar] [CrossRef]
- Nault, J.C.; Mallet, M.; Pilati, C.; Calderaro, J.; Bioulac-Sage, P.; Laurent, C.; Laurent, A.; Cherqui, D.; Balabaud, C.; Zucman-Rossi, J. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat. Commun. 2013, 4, 2218. [Google Scholar] [CrossRef] [Green Version]
- Johansson, G.; Andersson, D.; Filges, S.; Li, J.; Muth, A.; Godfrey, T.E.; Stahlberg, A. Considerations and quality controls when analyzing cell-free tumor DNA. Biomol. Detect. Quantif. 2019, 17, 100078. [Google Scholar] [CrossRef]
- World Medical, A. World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA 2013, 310, 2191–2194. [Google Scholar]
- Joo, I.; Kim, S.Y.; Kang, T.W.; Kim, Y.K.; Park, B.J.; Lee, Y.J.; Choi, J.I.; Lee, C.H.; Park, H.S.; Lee, K.; et al. Radiologic-Pathologic Correlation of Hepatobiliary Phase Hypointense Nodules without Arterial Phase Hyperenhancement at Gadoxetic Acid-enhanced MRI. A Multicenter Study. Radiology 2020, 296, 192275. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, P.; Yoon, S.; Kim, N.; Lee, S.; Ko, M.; Lee, H.; Kang, H.; Kim, J.; Lee, S. ChimerDB 2.0—A knowledgebase for fusion genes updated. Nucleic Acids Res. 2010, 38, D81–D85. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Variables | Total | |
|---|---|---|
| Patients | 40 | |
| Gender | male | 32 |
| female | 8 | |
| Age at inclusion | median ± SD (range) | 56 ± 10.41 (33–78) |
| <50 years | 8 | |
| ≥50 years | 32 | |
| Etiology | AIH | 2 |
| AC | 15 | |
| AC/HCV | 3 | |
| AC/AIH | 1 | |
| Viral | 3 | |
| NASH/PBC | 1 | |
| PBC/AIH | 2 | |
| PSC | 5 | |
| B–CS | 2 | |
| medication | 2 | |
| other | 3 | |
| BMI (kg/m2) | median (range) | 27.26 (18.62–41.52) |
| Child-Pugh score | A (5–6) | 12 |
| B (7–9) | 23 | |
| C (10–15) | 5 | |
| MELD score | <15 | 20 |
| ≥15 | 20 | |
| AFP (ng/mL) | <10 | 27 |
| ≥10 | 1 | |
| n.a. | 12 | |
| Diabetes | yes | 9 |
| no | 27 | |
| Portal vein thrombosis | yes | 4 |
| no | 36 | |
| (Chronic) renal failure | yes | 16 |
| no | 23 | |
| n.a. | 1 | |
| Ascites | yes | 21 |
| no | 19 | |
| Varices | yes | 35 |
| no | 5 | |
| Reasons for MRI | Transplantation evaluation | 1 |
| PSC | 5 | |
| Portal vein thrombosis | 2 | |
| Suboptimal ultrasound | 2 | |
| TIPS evaluation | 2 | |
| Cystic pancreatic lesion | 1 | |
| B–CS | 1 | |
| Post-transplant control | 1 |
| Gene | Chr:Position | Codon Change | AA Change | Variant Type |
|---|---|---|---|---|
| ABCB1 | Chr.7:87531302 | c.2677T > G | p.S893A | frameshift |
| c.2485T > G | p.S829A | |||
| AC055811.2 | Chr17:17216394 | c.119insC | - | frameshift |
| ASXL1 | Chr.20:32434638 | c.1919insG | p.G641fs | frameshift |
| c.1934insG | p.G646fs | |||
| AXIN2 | Chr.17:65536466 | c.1799delG | p.G600fs | frameshift |
| c.1994delG | p.G665fs | |||
| BAX | Chr.19:48955713 | c.121insG | p.E41fs | frameshift |
| c.69insG | p.R24fs | |||
| c.70insG | p.E24fs | |||
| BRAF | Chr.7:140777014 | c.1592G > T | p.W531L p.W138L | SNV |
| c.413G > T | ||||
| c. * 1042G > T | ||||
| BRCA2 | Chr13:32333283 | c.1813delA | p.I605fs | frameshift |
| BRCA2 | Chr13:32339421 | c.5073delA | p.K1691fs | frameshift |
| CD3EAP | Chr.19:45409478 | c.1516C > A | p.Q506K | SNV |
| c.1510C > A | p.Q504K | |||
| CHD2 | Chr15:93002203 | c.4173insA | p.Q1392fs | frameshift |
| CHEK2 | Chr22:28725242 | c. 5731G > A | - | SNV |
| c. * 424 + 1G > A | - | |||
| c.220 + 1G > A | - | |||
| c.474 + 1G > A | - | |||
| c.5731G > A | - | |||
| c.6 + 1G > A | - | |||
| c.444 + 1G > A | - | |||
| CYP2B6 | Chr19:41006936 | c.516G > T | p.Q172H | SNV |
| CYP2B6 | Chr19:41009358 | c.785A > G | p.K262R | SNV |
| ERCC1 | Chr19:45409478 | c.197G > T | - | SNV |
| FLCN | Chr17:17216394 | c.1285insC | p.H429fs | frameshift |
| HNF1A | Chr12:120994314 | c.685insC | p.Q229fs | frameshift |
| c.872insC | p.G292fs | |||
| MSH6 | Chr2:47803500 | c.165delC | p.F56fs | frameshift |
| c.3261delC | p.F1088fs | |||
| c. * 2608delC | - | |||
| c.2871delC | p.F958fs | |||
| c.2355delC | p.F786fs | |||
| MPL | Chr.43338725 | c.391 + 5G > C | - | SNV |
| c.370 + 5G > C | - | |||
| NBN | Chr8:89937066 | c. * 2067C > T | - | SNV |
| PTEN | Chr.10:87933154 | c.395G > A | p.G132D | SNV |
| SLCO1B1 | Chr12.21178615 | c.521T > C | p.V174A | SNV |
| WRN | Chr8:31058454 | c.15delA | p.K5fs | frameshift |
| Patients (n = 36) | Variant | % | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HNF1A | 28 | ||||||||||||||||||||||||||||||||||||
| BAX | 25 | ||||||||||||||||||||||||||||||||||||
| ASXL1 | 19 | ||||||||||||||||||||||||||||||||||||
| CHD2 | 11 | ||||||||||||||||||||||||||||||||||||
| CYP2B6 | 5 | ||||||||||||||||||||||||||||||||||||
| FLCN | 5 | ||||||||||||||||||||||||||||||||||||
| NBN | 5 | ||||||||||||||||||||||||||||||||||||
| ABCB1 | 3 | ||||||||||||||||||||||||||||||||||||
| AC055811.2 | 3 | ||||||||||||||||||||||||||||||||||||
| AXIN2 | 3 | ||||||||||||||||||||||||||||||||||||
| BRCA2 | 3 | ||||||||||||||||||||||||||||||||||||
| BRAF | 3 | ||||||||||||||||||||||||||||||||||||
| CD3EAP | 3 | ||||||||||||||||||||||||||||||||||||
| CHEK2 | 3 | ||||||||||||||||||||||||||||||||||||
| ERCC1 | 3 | ||||||||||||||||||||||||||||||||||||
| MSH6 | 3 | ||||||||||||||||||||||||||||||||||||
| MPL | 3 | ||||||||||||||||||||||||||||||||||||
| PTEN | 3 | ||||||||||||||||||||||||||||||||||||
| SLCO1B1 | 3 | ||||||||||||||||||||||||||||||||||||
| WRN | 3 | ||||||||||||||||||||||||||||||||||||
| Patient ID | Variants Detected by NGS | Second MRI Evaluation |
|---|---|---|
| none | no lesion | |
| none | no lesion | |
| none | no lesion | |
| none | no lesion | |
| none | no lesion | |
| none | no lesion | |
| none | no lesion | |
| none | no lesion | |
| HNF1A | no lesion | |
| HNF1A | no lesion | |
| HNF1A | no lesion | |
| HNF1A; BAX | no lesion | |
| BAX | no lesion | |
| CHD2; CYP2B6 | no lesion | |
| BRCA2; MSH6 | no lesion | |
| ASXL1; CHD2 | no lesion | |
| CYP2B6; WRN | no lesion | |
| ASXL1 | no lesion | |
| CD3EAP; ERCC1; NBN | no lesion | |
| AC055811.2; FLCN | no lesion | |
| BAX; ABCB1; MPL; SLCO1B1 | no lesion | |
| 92381 | BAX | early HCC |
| 92387 | HNF1A | early HCC |
| 92396 | HNF1A | HGDN |
| 92502 | BRAF; NBN; PTEN | HCC |
| 92505 | BAX; ASXL1; CHD2 | HCC |
| 92507 | HNF1A; ASXL1 | HCC |
| Patient’s ID | Variants | Second MRI Re-Evaluation |
|---|---|---|
| 92379 | none | no new lesion |
| 92381 | BAX | Early HCC (confirmed) HCC (newly discovered) |
| 92382 | AC055811.2; FLCN | no new lesion |
| 92389 | none | no new lesion |
| 92396 | HNF1A | HGDN (confirmed) |
| 92397 | HNF1A | no new lesion |
| 92500 | CYP2B6; WRN | no new lesion |
| 92502 | BRAF; NBN; PTEN | HCC (confirmed) HCC (newly discovered) |
| 92504 | ASXL1 | no new lesion |
| Sub-Cohort | cfDNA (ng/µL) | Sub-Cohort | cfDNA (ng/µL) | |
|---|---|---|---|---|
| Mean ± SD | Mean ± SD | p Values | ||
| LC (n = 36) | 42.27 ± 42.42 | HD (n = 7) | 8.46 ± 1.47 | <0.005 |
| LC (n = 30) | 39.58 ± 42.77 | HD (n = 7) | 8.46 ± 1.47 | <0.005 |
| Malignant lesion (n = 6) | 55.69 ± 37.83 | HD (n = 7) | 8.46 ± 1.47 | 0.007 |
| Malignant lesion (n = 6) | 55.69 ± 37.83 | LC (n = 30) | 39.58 ± 42.77 | 0.445 |
| Variable | Total (n = 36) | Low cfDNA (n = 21) | High cfDNA (n = 15) | p Value |
|---|---|---|---|---|
| Gender | 0.667 | |||
| female | 6 | 3 | 3 | |
| male | 30 | 18 | 12 | |
| Age | 0.077 | |||
| <60 years | 25 | 12 | 13 | |
| >60 years | 11 | 9 | 12 | |
| Etiology | 0.500 | |||
| AC positive | 18 | 12 | 6 | |
| AC negative | 18 | 9 | 9 | |
| BMI (kg/m2) | 0.499 | |||
| <25 | 14 | 7 | 7 | |
| >25 | 22 | 14 | 8 | |
| Child-Pugh score | 0.320 | |||
| ≤7 | 16 | 11 | 5 | |
| >7 | 20 | 10 | 10 | |
| MELD | 0.500 | |||
| <15 | 20 | 13 | 7 | |
| ≥15 | 16 | 8 | 8 | |
| AFP (ng/mL) | 0.024 | |||
| ≤7 | 25 | 12 | 13 | |
| >7 | 1 | 0 | 1 | |
| diabetes | 0.705 | |||
| no | 27 | 15 | 12 | |
| yes | 9 | 6 | 3 | |
| Portal vein thrombosis | 1.000 | |||
| no | 32 | 19 | 13 | |
| yes | 4 | 2 | 2 | |
| (Chronic) renal failure | 0.728 | |||
| no | 20 | 11 | 9 | |
| yes | 15 | 10 | 5 | |
| Ascites | 0.516 | |||
| no | 17 | 11 | 6 | |
| yes | 19 | 10 | 9 | |
| Varices | 1.000 | |||
| no | 4 | 2 | 2 | |
| yes | 32 | 19 | 13 |
| Variant | AC | HBV/HCV | NASH | PSC | B–CS | PBC/AIH | CPA | CPB | CPC |
|---|---|---|---|---|---|---|---|---|---|
| HNF1A | |||||||||
| BAX | |||||||||
| ASXL1 | |||||||||
| CHD2 | |||||||||
| CYP2B6 | |||||||||
| FLCN | |||||||||
| NBN | |||||||||
| AC055811.2 | |||||||||
| AXIN2 | |||||||||
| ABCB1 | |||||||||
| BRCA2 | |||||||||
| BRAF | |||||||||
| CD3EAP | |||||||||
| CHEK2 | |||||||||
| ERCC1 | |||||||||
| MSH6 | |||||||||
| MPL | |||||||||
| PTEN | |||||||||
| SLCO1B1 | |||||||||
| WRN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alunni-Fabbroni, M.; Weber, S.; Öcal, O.; Seidensticker, M.; Mayerle, J.; Malfertheiner, P.; Ricke, J. Circulating Cell-Free DNA Combined to Magnetic Resonance Imaging for Early Detection of HCC in Patients with Liver Cirrhosis. Cancers 2021, 13, 521. https://doi.org/10.3390/cancers13030521
Alunni-Fabbroni M, Weber S, Öcal O, Seidensticker M, Mayerle J, Malfertheiner P, Ricke J. Circulating Cell-Free DNA Combined to Magnetic Resonance Imaging for Early Detection of HCC in Patients with Liver Cirrhosis. Cancers. 2021; 13(3):521. https://doi.org/10.3390/cancers13030521
Chicago/Turabian StyleAlunni-Fabbroni, Marianna, Sabine Weber, Osman Öcal, Max Seidensticker, Julia Mayerle, Peter Malfertheiner, and Jens Ricke. 2021. "Circulating Cell-Free DNA Combined to Magnetic Resonance Imaging for Early Detection of HCC in Patients with Liver Cirrhosis" Cancers 13, no. 3: 521. https://doi.org/10.3390/cancers13030521
APA StyleAlunni-Fabbroni, M., Weber, S., Öcal, O., Seidensticker, M., Mayerle, J., Malfertheiner, P., & Ricke, J. (2021). Circulating Cell-Free DNA Combined to Magnetic Resonance Imaging for Early Detection of HCC in Patients with Liver Cirrhosis. Cancers, 13(3), 521. https://doi.org/10.3390/cancers13030521