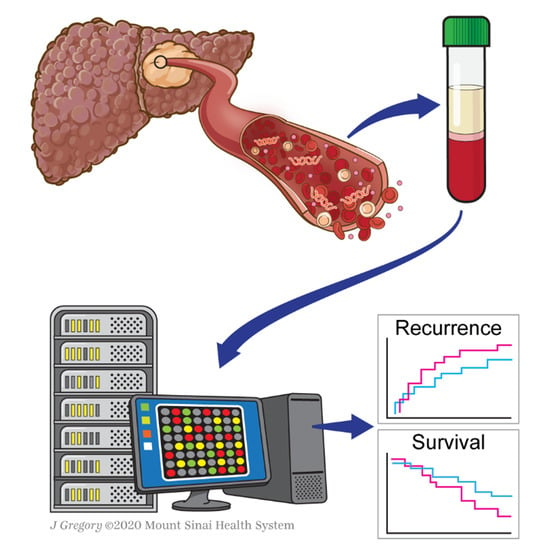
The Role of Liquid Biopsy in Hepatocellular Carcinoma Prognostication
Abstract
:Simple Summary
Abstract
1. Background
1.1. Circulating Tumor DNA (ctDNA)
1.1.1. Copy Number Variations (CNVs)
1.1.2. Mutations
1.1.3. DNA Methylation Changes
1.2. Circulating Free RNAs (cfRNAs)
1.2.1. Micro-RNAs (miRNAs)
1.2.2. Messenger RNAs (mRNAs)
1.3. Extracellular Vesicles (EVs): Exosomes
1.4. Circulating Tumor Cells (CTCs)
2. Challenges and Future Perspectives
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Projections of Mortality and Causes of Death, 2016 to 2060. Available online: http://www.who.int/healthinfo/global_burden_disease/projections/en/ (accessed on 14 October 2018).
- Llovet, J.M.; Bru, C.; Bruix, J. Prognosis of hepatocellular carcinoma: The BCLC staging classification. Semin. Liver. Dis. 1999, 19, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Bruix, J.; Sherman, M.; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: An update. Hepatology 2011, 53, 1020–1022. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Peng, W.; Liu, X.; Li, C.; Li, X.; Wen, T.F. Post-treatment alpha-fetoprotein response predicts prognosis of patients with hepatocellular carcinoma: A meta-analysis. Medicine (Baltimore) 2019, 98, e16557. [Google Scholar] [CrossRef] [PubMed]
- Labgaa, I.; Torrecilla, S.; Martinez-Quetglas, I.; Sia, D. Genetics of Hepatocellular Carcinoma: Risk Stratification, Clinical Outcome, and Implications for Therapy. Digest. Disease Interv. 2017, 01, 055–065. [Google Scholar] [CrossRef]
- Goossens, N.; Labgaa, I.; Villanueva, A. Nontumor Prognostic Factors in Hepatocellular Carcinoma. In Hepatocellular Carcinoma. Current Clinical Oncology; Carr, B.I., Ed.; Springer: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Hoshida, Y.; Villanueva, A.; Kobayashi, M.; Peix, J.; Chiang, D.Y.; Camargo, A.; Gupta, S.; Moore, J.; Wrobel, M.J.; Lerner, J.; et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 1995–2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villanueva, A.; Hoshida, Y.; Battiston, C.; Tovar, V.; Sia, D.; Alsinet, C.; Cornella, H.; Liberzon, A.; Kobayashi, M.; Kumada, H.; et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology 2011, 140, 1501–1512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [Green Version]
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abecassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018, 67, 358–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, M.A.; Hegab, B.; Hyde, C.; Guo, B.; Buckels, J.A.; Mirza, D.F. Needle track seeding following biopsy of liver lesions in the diagnosis of hepatocellular cancer: A systematic review and meta-analysis. Gut 2008, 57, 1592–1596. [Google Scholar] [CrossRef]
- Labgaa, I.; Villanueva, A. Liquid biopsy in liver cancer. Discov. Med. 2015, 19, 263–273. [Google Scholar]
- von Felden, J.; Garcia-Lezana, T.; Schulze, K.; Losic, B.; Villanueva, A. Liquid biopsy in the clinical management of hepatocellular carcinoma. Gut 2020, 69, 2025–2034. [Google Scholar] [CrossRef]
- Labgaa, I.; Craig, A.J.; Villanueva, A. Diagnostic and Prognostic Performance of Liquid Biopsy in Hepatocellular Carcinoma. In Liquid Biopsy in Cancer Patients. Current Clinical Pathology; Giordano, A., Rolfo, C., Russo, A., Eds.; Humana Press: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [Green Version]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346. [Google Scholar] [CrossRef] [Green Version]
- Tie, J.; Cohen, J.D.; Wang, Y.; Christie, M.; Simons, K.; Lee, M.; Wong, R.; Kosmider, S.; Ananda, S.; McKendrick, J.; et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019. [Google Scholar] [CrossRef]
- Lee, B.; Lipton, L.; Cohen, J.; Tie, J.; Javed, A.A.; Li, L.; Goldstein, D.; Burge, M.; Cooray, P.; Nagrial, A.; et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann. Oncol. 2019, 30, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Parikh, A.R.; Leshchiner, I.; Elagina, L.; Goyal, L.; Levovitz, C.; Siravegna, G.; Livitz, D.; Rhrissorrakrai, K.; Martin, E.E.; Van Seventer, E.E.; et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 2019, 25, 1415–1421. [Google Scholar] [CrossRef]
- Russo, M.; Misale, S.; Wei, G.; Siravegna, G.; Crisafulli, G.; Lazzari, L.; Corti, G.; Rospo, G.; Novara, L.; Mussolin, B.; et al. Acquired Resistance to the TRK Inhibitor Entrectinib in Colorectal Cancer. Cancer Discov. 2016, 6, 36–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, M.; Siravegna, G.; Blaszkowsky, L.S.; Corti, G.; Crisafulli, G.; Ahronian, L.G.; Mussolin, B.; Kwak, E.L.; Buscarino, M.; Lazzari, L.; et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016, 6, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef] [Green Version]
- De Rubis, G.; Rajeev Krishnan, S.; Bebawy, M. Liquid Biopsies in Cancer Diagnosis, Monitoring, and Prognosis. Trends Pharmacol. Sci. 2019, 40, 172–186. [Google Scholar] [CrossRef]
- Jiang, P.; Chan, C.W.; Chan, K.C.; Cheng, S.H.; Wong, J.; Wong, V.W.; Wong, G.L.; Chan, S.L.; Mok, T.S.; Chan, H.L.; et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl. Acad. Sci. USA 2015, 112, E1317–E1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labgaa, I.; Villacorta-Martin, C.; D’Avola, D.; Craig, A.J.; von Felden, J.; Martins-Filho, S.N.; Sia, D.; Stueck, A.; Ward, S.C.; Fiel, M.I.; et al. A pilot study of ultra-deep targeted sequencing of plasma DNA identifies driver mutations in hepatocellular carcinoma. Oncogene 2018, 37, 3740–3752. [Google Scholar] [CrossRef]
- Cai, Z.; Chen, G.; Zeng, Y.; Dong, X.; Li, Z.; Huang, Y.; Xin, F.; Qiu, L.; Xu, H.; Zhang, W.; et al. Comprehensive Liquid Profiling of Circulating Tumor DNA and Protein Biomarkers in Long-Term Follow-Up Patients with Hepatocellular Carcinoma. Clin. Cancer Res. 2019, 25, 5284–5294. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.R.; Kong, S.Y.; Im, H.S.; Kim, H.J.; Kim, M.K.; Yoon, K.A.; Cho, E.H.; Jang, J.H.; Lee, J.; Kang, J.; et al. Genome-wide copy number alteration and VEGFA amplification of circulating cell-free DNA as a biomarker in advanced hepatocellular carcinoma patients treated with Sorafenib. BMC Cancer 2019, 19, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, A.; Fujimoto, A.; Yamamoto, Y.; Akamatsu, S.; Hiraga, N.; Imamura, M.; Kawaoka, T.; Tsuge, M.; Abe, H.; Hayes, C.N.; et al. Circulating Tumor DNA Analysis for Liver Cancers and Its Usefulness as a Liquid Biopsy. Cell Mol. Gastroenterol. Hepatol. 2015, 1, 516–534. [Google Scholar] [CrossRef] [Green Version]
- Liao, W.; Yang, H.; Xu, H.; Wang, Y.; Ge, P.; Ren, J.; Xu, W.; Lu, X.; Sang, X.; Zhong, S.; et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget 2016, 7, 40481–40490. [Google Scholar] [CrossRef] [Green Version]
- Hann, H.W.; Jain, S.; Park, G.; Steffen, J.D.; Song, W.; Su, Y.H. Detection of urine DNA markers for monitoring recurrent hepatocellular carcinoma. Hepatoma Res. 2017, 3, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Jiao, J.; Watt, G.P.; Stevenson, H.L.; Calderone, T.L.; Fisher-Hoch, S.P.; Ye, Y.; Wu, X.; Vierling, J.M.; Beretta, L. Telomerase reverse transcriptase mutations in plasma DNA in patients with hepatocellular carcinoma or cirrhosis: Prevalence and risk factors. Hepatol. Commun. 2018, 2, 718–731. [Google Scholar] [CrossRef]
- Oversoe, S.K.; Clement, M.S.; Pedersen, M.H.; Weber, B.; Aagaard, N.K.; Villadsen, G.E.; Gronbaek, H.; Hamilton-Dutoit, S.J.; Sorensen, B.S.; Kelsen, J. TERT promoter mutated circulating tumor DNA as a biomarker for prognosis in hepatocellular carcinoma. Scand. J. Gastroenterol. 2020, 55, 1433–1440. [Google Scholar] [CrossRef]
- Kim, S.S.; Eun, J.W.; Choi, J.H.; Woo, H.G.; Cho, H.J.; Ahn, H.R.; Suh, C.W.; Baek, G.O.; Cho, S.W.; Cheong, J.Y. MLH1 single-nucleotide variant in circulating tumor DNA predicts overall survival of patients with hepatocellular carcinoma. Sci. Rep. 2020, 10, 17862. [Google Scholar] [CrossRef]
- Hirai, M.; Kinugasa, H.; Nouso, K.; Yamamoto, S.; Terasawa, H.; Onishi, Y.; Oyama, A.; Adachi, T.; Wada, N.; Sakata, M.; et al. Prediction of the prognosis of advanced hepatocellular carcinoma by TERT promoter mutations in circulating tumor DNA. J. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Li, S.F.; Wang, J.L.; Zhang, T.; Zhang, S.; Chen, H.T.; Xiao, Q.Y.; Ren, W.H.; Liu, C.; Peng, B.; et al. TP53 R249S mutation detected in circulating tumour DNA is associated with Prognosis of hepatocellular carcinoma patients with or without hepatectomy. Liver Int. 2020, 40, 2834–2847. [Google Scholar] [CrossRef]
- von Felden, J.; Craig, A.J.; Garcia-Lezana, T.; Labgaa, I.; Haber, P.K.; D’Avola, D.; Asgharpour, A.; Dieterich, D.; Bonaccorso, A.; Torres-Martin, M.; et al. Mutations in circulating tumor DNA predict primary resistance to systemic therapies in advanced hepatocellular carcinoma. Oncogene 2020. [Google Scholar] [CrossRef]
- Huang, Z.H.; Hu, Y.; Hua, D.; Wu, Y.Y.; Song, M.X.; Cheng, Z.H. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp. Mol. Pathol. 2011, 91, 702–707. [Google Scholar] [CrossRef]
- Xu, R.H.; Wei, W.; Krawczyk, M.; Wang, W.; Luo, H.; Flagg, K.; Yi, S.; Shi, W.; Quan, Q.; Li, K.; et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017, 16, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.Y.; Chen, S.Y.; Peng, H.L.; Kan, P.Y.; Chang, W.C.; Yen, C.J. Cell-free methylation markers with diagnostic and prognostic potential in hepatocellular carcinoma. Oncotarget 2017, 8, 6406–6418. [Google Scholar] [CrossRef] [Green Version]
- Yeh, C.C.; Goyal, A.; Shen, J.; Wu, H.C.; Strauss, J.A.; Wang, Q.; Gurvich, I.; Safyan, R.A.; Manji, G.A.; Gamble, M.V.; et al. Global Level of Plasma DNA Methylation is Associated with Overall Survival in Patients with Hepatocellular Carcinoma. Ann. Surg. Oncol. 2017, 24, 3788–3795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Qiao, C.Y.; Gao, S.; Fan, Y.C.; Chen, L.Y.; Wang, K. Circulating cell-free DNA of methylated insulin-like growth factor-binding protein 7 predicts a poor prognosis in hepatitis B virus-associated hepatocellular carcinoma after hepatectomy. Free Radic. Res. 2018, 52, 455–464. [Google Scholar] [CrossRef]
- Chen, M.M.; Zhao, R.C.; Chen, K.F.; Huang, Y.; Liu, Z.J.; Wei, Y.G.; Jian, Y.; Sun, A.M.; Qin, L.; Li, B.; et al. Hypomethylation of CTCFL promoters as a noninvasive biomarker in plasma from patients with hepatocellular carcinoma. Neoplasma 2020, 67, 909–915. [Google Scholar] [CrossRef]
- Chiang, D.Y.; Villanueva, A.; Hoshida, Y.; Peix, J.; Newell, P.; Minguez, B.; LeBlanc, A.C.; Donovan, D.J.; Thung, S.N.; Sole, M.; et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008, 68, 6779–6788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulze, K.; Imbeaud, S.; Letouze, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef]
- Totoki, Y.; Tatsuno, K.; Covington, K.R.; Ueda, H.; Creighton, C.J.; Kato, M.; Tsuji, S.; Donehower, L.A.; Slagle, B.L.; Nakamura, H.; et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014, 46, 1267–1273. [Google Scholar] [CrossRef]
- Hernandez-Meza, G.; von Felden, J.; Gonzalez-Kozlova, E.E.; Garcia-Lezana, T.; Peix, J.; Portela, A.; Craig, A.J.; Sayols, S.; Schwartz, M.; Losic, B.; et al. DNA methylation profiling of human hepatocarcinogenesis. Hepatology 2020. [Google Scholar] [CrossRef]
- Villanueva, A.; Portela, A.; Sayols, S.; Battiston, C.; Hoshida, Y.; Mendez-Gonzalez, J.; Imbeaud, S.; Letouze, E.; Hernandez-Gea, V.; Cornella, H.; et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology 2015, 61, 1945–1956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koberle, V.; Kronenberger, B.; Pleli, T.; Trojan, J.; Imelmann, E.; Peveling-Oberhag, J.; Welker, M.W.; Elhendawy, M.; Zeuzem, S.; Piiper, A.; et al. Serum microRNA-1 and microRNA-122 are prognostic markers in patients with hepatocellular carcinoma. Eur. J. Cancer 2013, 49, 3442–3449. [Google Scholar] [CrossRef]
- Xu, Y.; Bu, X.; Dai, C.; Shang, C. High serum microRNA-122 level is independently associated with higher overall survival rate in hepatocellular carcinoma patients. Tumour. Biol. 2015, 36, 4773–4776. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Kim, S.S.; Nam, J.S.; Kim, J.K.; Lee, J.H.; Kim, B.; Wang, H.J.; Kim, B.W.; Lee, J.D.; Kang, D.Y.; et al. Low levels of circulating microRNA-26a/29a as poor prognostic markers in patients with hepatocellular carcinoma who underwent curative treatment. Clin. Res. Hepatol. Gastroenterol. 2017, 41, 181–189. [Google Scholar] [CrossRef]
- Ning, S.; Liu, H.; Gao, B.; Wei, W.; Yang, A.; Li, J.; Zhang, L. miR-155, miR-96 and miR-99a as potential diagnostic and prognostic tools for the clinical management of hepatocellular carcinoma. Oncol. Lett. 2019, 18, 3381–3387. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Wong, Y.S.; Goh, B.K.P.; Chan, C.Y.; Cheow, P.C.; Chow, P.K.H.; Lim, T.K.H.; Goh, G.B.B.; Krishnamoorthy, T.L.; Kumar, R.; et al. Circulating microRNAs as Potential Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. Sci. Rep. 2019, 9, 10464. [Google Scholar] [CrossRef] [Green Version]
- Loosen, S.H.; Wirtz, T.H.; Roy, S.; Vucur, M.; Castoldi, M.; Schneider, A.T.; Koppe, C.; Ulmer, T.F.; Roeth, A.A.; Bednarsch, J.; et al. Circulating levels of microRNA193a-5p predict outcome in early stage hepatocellular carcinoma. PLoS ONE 2020, 15, e0239386. [Google Scholar] [CrossRef]
- Pratedrat, P.; Chuaypen, N.; Nimsamer, P.; Payungporn, S.; Pinjaroen, N.; Sirichindakul, B.; Tangkijvanich, P. Diagnostic and prognostic roles of circulating miRNA-223-3p in hepatitis B virus-related hepatocellular carcinoma. PLoS ONE 2020, 15, e0232211. [Google Scholar] [CrossRef] [Green Version]
- Jeng, K.S.; Sheen, I.S.; Wang, Y.C.; Gu, S.L.; Chu, C.M.; Shih, S.C.; Wang, P.C.; Chang, W.H.; Wang, H.Y. Prognostic significance of preoperative circulating vascular endothelial growth factor messenger RNA expression in resectable hepatocellular carcinoma: A prospective study. World J. Gastroenterol. 2004, 10, 643–648. [Google Scholar] [CrossRef]
- Morimoto, O.; Nagano, H.; Miyamoto, A.; Fujiwara, Y.; Kondo, M.; Yamamoto, T.; Ota, H.; Nakamura, M.; Wada, H.; Damdinsuren, B.; et al. Association between recurrence of hepatocellular carcinoma and alpha-fetoprotein messenger RNA levels in peripheral blood. Surg. Today 2005, 35, 1033–1041. [Google Scholar] [CrossRef]
- Kong, S.Y.; Park, J.W.; Kim, J.O.; Lee, N.O.; Lee, J.A.; Park, K.W.; Hong, E.K.; Kim, C.M. Alpha-fetoprotein and human telomerase reverse transcriptase mRNA levels in peripheral blood of patients with hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2009, 135, 1091–1098. [Google Scholar] [CrossRef] [PubMed]
- Sugimachi, K.; Matsumura, T.; Hirata, H.; Uchi, R.; Ueda, M.; Ueo, H.; Shinden, Y.; Iguchi, T.; Eguchi, H.; Shirabe, K.; et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. Cancer 2015, 112, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Wu, J.; Wu, J.; Ji, A.; Qiang, G.; Jiang, Y.; Jiang, C.; Ding, Y. Exosomal miR-665 as a novel minimally invasive biomarker for hepatocellular carcinoma diagnosis and prognosis. Oncotarget 2017, 8, 80666–80678. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.R.; Kim, G.; Tak, W.Y.; Jang, S.Y.; Kweon, Y.O.; Park, J.G.; Lee, H.W.; Han, Y.S.; Chun, J.M.; Park, S.Y.; et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int. J. Cancer 2019, 144, 1444–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, M.; Jiang, Y.; Yang, L.; Yan, S.; Wang, Y.G.; Lu, X.J. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J. Cell Biochem. 2018, 119, 4711–4716. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Cao, J.; Chen, L.; Xi, X.; Wang, S.; Zhu, Y.; Yang, L.; Ma, L.; Wang, D.; Yin, J.; et al. Noncoding RNAs Serve as Diagnosis and Prognosis Biomarkers for Hepatocellular Carcinoma. Clin. Chem. 2019, 65, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Liu, F.; Gui, R. High expression of circulating exosomal circAKT3 is associated with higher recurrence in HCC patients undergoing surgical treatment. Surg. Oncol. 2020, 33, 276–281. [Google Scholar] [CrossRef]
- Hao, X.; Xin, R.; Dong, W. Decreased serum exosomal miR-320a expression is an unfavorable prognostic factor in patients with hepatocellular carcinoma. J. Int. Med. Res. 2020, 48, 300060519896144. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.J.; Chong, Y.; Guo, Z.W.; Xie, C.; Yang, X.J.; Zhang, Q.; Li, S.P.; Xiong, Y.; Yuan, Y.; Min, J.; et al. A serum microRNA classifier for early detection of hepatocellular carcinoma: A multicentre, retrospective, longitudinal biomarker identification study with a nested case-control study. Lancet Oncol. 2015, 16, 804–815. [Google Scholar] [CrossRef]
- Maisano, D.; Mimmi, S.; Russo, R.; Fioravanti, A.; Fiume, G.; Vecchio, E.; Nistico, N.; Quinto, I.; Iaccino, E. Uncovering the Exosomes Diversity: A Window of Opportunity for Tumor Progression Monitoring. Pharmaceuticals 2020, 13, 180. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e1018. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Iaccino, E.; Mimmi, S.; Dattilo, V.; Marino, F.; Candeloro, P.; Di Loria, A.; Marimpietri, D.; Pisano, A.; Albano, F.; Vecchio, E.; et al. Monitoring multiple myeloma by idiotype-specific peptide binders of tumor-derived exosomes. Mol. Cancer 2017, 16, 159. [Google Scholar] [CrossRef] [PubMed]
- Nistico, N.; Maisano, D.; Iaccino, E.; Vecchio, E.; Fiume, G.; Rotundo, S.; Quinto, I.; Mimmi, S. Role of Chronic Lymphocytic Leukemia (CLL)-Derived Exosomes in Tumor Progression and Survival. Pharmaceuticals 2020, 13, 244. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.F.; Xu, Y.; Yang, X.R.; Guo, W.; Zhang, X.; Qiu, S.J.; Shi, R.Y.; Hu, B.; Zhou, J.; Fan, J. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology 2013, 57, 1458–1468. [Google Scholar] [CrossRef]
- D’Avola, D.; Villacorta-Martin, C.; Martins-Filho, S.N.; Craig, A.; Labgaa, I.; von Felden, J.; Kimaada, A.; Bonaccorso, A.; Tabrizian, P.; Hartmann, B.M.; et al. High-density single cell mRNA sequencing to characterize circulating tumor cells in hepatocellular carcinoma. Sci. Rep. 2018, 8, 11570. [Google Scholar] [CrossRef] [PubMed]
- Vona, G.; Estepa, L.; Beroud, C.; Damotte, D.; Capron, F.; Nalpas, B.; Mineur, A.; Franco, D.; Lacour, B.; Pol, S.; et al. Impact of cytomorphological detection of circulating tumor cells in patients with liver cancer. Hepatology 2004, 39, 792–797. [Google Scholar] [CrossRef]
- Xu, W.; Cao, L.; Chen, L.; Li, J.; Zhang, X.F.; Qian, H.H.; Kang, X.Y.; Zhang, Y.; Liao, J.; Shi, L.H.; et al. Isolation of circulating tumor cells in patients with hepatocellular carcinoma using a novel cell separation strategy. Clin. Cancer Res. 2011, 17, 3783–3793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, S.T.; Yang, Z.F.; Ho, D.W.; Ng, M.N.; Yu, W.C.; Wong, J. Prediction of posthepatectomy recurrence of hepatocellular carcinoma by circulating cancer stem cells: A prospective study. Ann. Surg. 2011, 254, 569–576. [Google Scholar] [CrossRef]
- Cheng, S.W.; Tsai, H.W.; Lin, Y.J.; Cheng, P.N.; Chang, Y.C.; Yen, C.J.; Huang, H.P.; Chuang, Y.P.; Chang, T.T.; Lee, C.T.; et al. Lin28B is an oncofetal circulating cancer stem cell-like marker associated with recurrence of hepatocellular carcinoma. PLoS ONE 2013, 8, e80053. [Google Scholar] [CrossRef]
- Liu, S.; Li, N.; Yu, X.; Xiao, X.; Cheng, K.; Hu, J.; Wang, J.; Zhang, D.; Cheng, S.; Liu, S. Expression of intercellular adhesion molecule 1 by hepatocellular carcinoma stem cells and circulating tumor cells. Gastroenterology 2013, 144, 1031–1041.e1010. [Google Scholar] [CrossRef] [Green Version]
- Schulze, K.; Gasch, C.; Staufer, K.; Nashan, B.; Lohse, A.W.; Pantel, K.; Riethdorf, S.; Wege, H. Presence of EpCAM-positive circulating tumor cells as biomarker for systemic disease strongly correlates to survival in patients with hepatocellular carcinoma. Int. J. Cancer 2013, 133, 2165–2171. [Google Scholar] [CrossRef]
- Guo, W.; Yang, X.R.; Sun, Y.F.; Shen, M.N.; Ma, X.L.; Wu, J.; Zhang, C.Y.; Zhou, Y.; Xu, Y.; Hu, B.; et al. Clinical significance of EpCAM mRNA-positive circulating tumor cells in hepatocellular carcinoma by an optimized negative enrichment and qRT-PCR-based platform. Clin. Cancer Res. 2014, 20, 4794–4805. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Shi, L.; Zhang, X.; Sun, B.; Yang, Y.; Ge, N.; Liu, H.; Yang, X.; Chen, L.; Qian, H.; et al. pERK/pAkt phenotyping in circulating tumor cells as a biomarker for sorafenib efficacy in patients with advanced hepatocellular carcinoma. Oncotarget 2016, 7, 2646–2659. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Niu, X.; Zou, L.; Li, L.; Li, S.; Han, J.; Zhang, P.; Song, J.; Xiao, F. AFP mRNA level in enriched circulating tumor cells from hepatocellular carcinoma patient blood samples is a pivotal predictive marker for metastasis. Cancer Lett. 2016, 378, 33–37. [Google Scholar] [CrossRef] [Green Version]
- Ogle, L.F.; Orr, J.G.; Willoughby, C.E.; Hutton, C.; McPherson, S.; Plummer, R.; Boddy, A.V.; Curtin, N.J.; Jamieson, D.; Reeves, H.L. Imagestream detection and characterisation of circulating tumour cells—A liquid biopsy for hepatocellular carcinoma? J. Hepatol. 2016, 65, 305–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- von Felden, J.; Schulze, K.; Krech, T.; Ewald, F.; Nashan, B.; Pantel, K.; Lohse, A.W.; Riethdorf, S.; Wege, H. Circulating tumor cells as liquid biomarker for high HCC recurrence risk after curative liver resection. Oncotarget 2017, 8, 89978–89987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Li, Y.; Xu, J.; Zhang, A.; Wang, X.; Tang, R.; Zhang, X.; Yin, H.; Liu, M.; Wang, D.D.; et al. Quantified postsurgical small cell size CTCs and EpCAM(+) circulating tumor stem cells with cytogenetic abnormalities in hepatocellular carcinoma patients determine cancer relapse. Cancer Lett. 2018, 412, 99–107. [Google Scholar] [CrossRef]
- Hao, S.; Chen, S.; Tu, C.; Huang, T. Anterior Approach to Improve the Prognosis in HCC Patients Via Decreasing Dissemination of EpCAM(+) Circulating Tumor Cells. J. Gastrointest Surg. 2017, 21, 1112–1120. [Google Scholar] [CrossRef]
- Sun, Y.F.; Guo, W.; Xu, Y.; Shi, Y.H.; Gong, Z.J.; Ji, Y.; Du, M.; Zhang, X.; Hu, B.; Huang, A.; et al. Circulating Tumor Cells from Different Vascular Sites Exhibit Spatial Heterogeneity in Epithelial and Mesenchymal Composition and Distinct Clinical Significance in Hepatocellular Carcinoma. Clin. Cancer Res. 2018, 24, 547–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, W.; Sun, Y.F.; Shen, M.N.; Ma, X.L.; Wu, J.; Zhang, C.Y.; Zhou, Y.; Xu, Y.; Hu, B.; Zhang, M.; et al. Circulating Tumor Cells with Stem-Like Phenotypes for Diagnosis, Prognosis, and Therapeutic Response Evaluation in Hepatocellular Carcinoma. Clin. Cancer Res. 2018, 24, 2203–2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, L.N.; Xiang, B.D.; Wu, F.X.; Ye, J.Z.; Zhong, J.H.; Wang, Y.Y.; Chen, Y.Y.; Chen, Z.S.; Ma, L.; Chen, J.; et al. Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients with Hepatocellular Carcinoma. Cancer Res. 2018, 78, 4731–4744. [Google Scholar] [CrossRef] [Green Version]
- Court, C.M.; Hou, S.; Winograd, P.; Segel, N.H.; Li, Q.W.; Zhu, Y.; Sadeghi, S.; Finn, R.S.; Ganapathy, E.; Song, M.; et al. A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellular carcinoma. Liver Transpl. 2018, 24, 946–960. [Google Scholar] [CrossRef]
- Yu, J.J.; Xiao, W.; Dong, S.L.; Liang, H.F.; Zhang, Z.W.; Zhang, B.X.; Huang, Z.Y.; Chen, Y.F.; Zhang, W.G.; Luo, H.P.; et al. Effect of surgical liver resection on circulating tumor cells in patients with hepatocellular carcinoma. BMC Cancer 2018, 18, 835. [Google Scholar] [CrossRef]
- Ha, Y.; Kim, T.H.; Shim, J.E.; Yoon, S.; Jun, M.J.; Cho, Y.H.; Lee, H.C. Circulating tumor cells are associated with poor outcomes in early-stage hepatocellular carcinoma: A prospective study. Hepatol. Int. 2019, 13, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Hamaoka, M.; Kobayashi, T.; Tanaka, Y.; Mashima, H.; Ohdan, H. Clinical significance of glypican-3-positive circulating tumor cells of hepatocellular carcinoma patients: A prospective study. PLoS ONE 2019, 14, e0217586. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Lin, X.; Chen, C.; Chen, Y.; Zhao, Q.; Wu, L.; Wang, D.; Ma, Y.; Ju, W.; Chen, M.; et al. Analysis of preoperative circulating tumor cells for recurrence in patients with hepatocellular carcinoma after liver transplantation. Ann. Transl. Med. 2020, 8, 1067. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, Z.; Zhou, H.; Leng, C.; Hou, B.; Zhou, C.; Hu, X.; Wang, J.; Chen, X. Preoperative circulating tumor cells to predict microvascular invasion and dynamical detection indicate the prognosis of hepatocellular carcinoma. BMC Cancer 2020, 20, 1047. [Google Scholar] [CrossRef]
- Winograd, P.; Hou, S.; Court, C.M.; Lee, Y.T.; Chen, P.J.; Zhu, Y.; Sadeghi, S.; Finn, R.S.; Teng, P.C.; Wang, J.J.; et al. Hepatocellular Carcinoma-Circulating Tumor Cells Expressing PD-L1 Are Prognostic and Potentially Associated With Response to Checkpoint Inhibitors. Hepatol. Commun. 2020, 4, 1527–1540. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.X.; Sun, Y.F.; Zhou, K.Q.; Cheng, J.W.; Hu, B.; Guo, W.; Yin, Y.; Huang, J.F.; Zhou, J.; Fan, J.; et al. Circulating tumor cells are an indicator for the administration of adjuvant transarterial chemoembolization in hepatocellular carcinoma: A single-center, retrospective, propensity-matched study. Clin. Transl. Med. 2020, 10, e137. [Google Scholar] [CrossRef]
- Wang, P.X.; Xu, Y.; Sun, Y.F.; Cheng, J.W.; Zhou, K.Q.; Wu, S.Y.; Hu, B.; Zhang, Z.F.; Guo, W.; Cao, Y.; et al. Detection of circulating tumour cells enables early recurrence prediction in hepatocellular carcinoma patients undergoing liver transplantation. Liver Int. 2020. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Forgues, M.; Wang, W.; Kim, J.W.; Ye, Q.; Jia, H.; Budhu, A.; Zanetti, K.A.; Chen, Y.; Qin, L.X.; et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008, 68, 1451–1461. [Google Scholar] [CrossRef] [Green Version]
- Dent, B.M.; Ogle, L.F.; O’Donnell, R.L.; Hayes, N.; Malik, U.; Curtin, N.J.; Boddy, A.V.; Plummer, E.R.; Edmondson, R.J.; Reeves, H.L.; et al. High-resolution imaging for the detection and characterisation of circulating tumour cells from patients with oesophageal, hepatocellular, thyroid and ovarian cancers. Int. J. Cancer 2016, 138, 206–216. [Google Scholar] [CrossRef] [Green Version]
- Toso, C.; Mentha, G.; Majno, P. Liver transplantation for hepatocellular carcinoma: Five steps to prevent recurrence. Am. J. Transplant. 2011, 11, 2031–2035. [Google Scholar] [CrossRef] [PubMed]
- Cui, K.; Ou, Y.; Shen, Y.; Li, S.; Sun, Z. Clinical value of circulating tumor cells for the diagnosis and prognosis of hepatocellular carcinoma (HCC): A systematic review and meta-analysis. Medicine (Baltimore) 2020, 99, e22242. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Fan, Z.; Wu, X.; Xu, M.; Jiang, J.; Tan, C.; Wu, W.; Wei, X.; Zhou, J. Circulating tumor cells are correlated with disease progression and treatment response in an orthotopic hepatocellular carcinoma model. Cytometry A 2015, 87, 1020–1028. [Google Scholar] [CrossRef]
- Gasparello, J.; Allegretti, M.; Tremante, E.; Fabbri, E.; Amoreo, C.A.; Romania, P.; Melucci, E.; Messana, K.; Borgatti, M.; Giacomini, P.; et al. Liquid biopsy in mice bearing colorectal carcinoma xenografts: Gateways regulating the levels of circulating tumor DNA (ctDNA) and miRNA (ctmiRNA). J. Exp. Clin. Cancer Res. 2018, 37, 124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Losic, B.; Craig, A.J.; Villacorta-Martin, C.; Martins-Filho, S.N.; Akers, N.; Chen, X.; Ahsen, M.E.; von Felden, J.; Labgaa, I.; D’Avola, D.; et al. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat. Commun. 2020, 11, 291. [Google Scholar] [CrossRef]


| Number of Patients | Treatment | Biomarkers | Technique | Main Finding | [Ref.] |
|---|---|---|---|---|---|
| CNV | |||||
| 34 HCC | Surgery | ctDNA (harboring SNV or CNV) | Targeted-sequencing and low coverage whole-genome sequencing | ctDNA can detect minimal residual disease (MRD) and predict survival | [28] |
| 151 HCC; 14 healthy controls | Sorafenib | VEGFA amplification | Whole-genome sequencing | High concentration of cell-free DNA (cfDNA) was associated with poor outcomes but VEGFA ratio was not a prognostic factor. | [29] |
| Mutations | |||||
| 46 HCC | Surgery Transplant | ctDNA | Targeted-sequencing and exome-sequencing | Detection of ctDNA was associated with increased recurrence | [30] |
| 41 HCC; 10 controls | Surgery | TERT, TP53 and CTNNB1 | Targeted-sequencing | Detection of ctDNA predicted shorter recurrence-free survival | [31] |
| 10 HCC | Surgery TACE RFA | Methylation of GSTP1 and RASSF1A or TP53 mutation | Methylation-specific PCR and sanger sequencing | Detecting ctDNA in urine was feasible and predicted recurrence | [32] |
| 218 HCC; 81 cirrhotic | NA | TERT promoter mutation (C228T and C250T) | Droplet digital PCR (ddPCR) and sanger sequencing | TERT promoter mutation can be used as an early biomarker of HCC and is associated with survival | [33] |
| 34 HCC | Surgery | ctDNA (harboring SNV or CNV) | Targeted-sequencing and low coverage whole-genome sequencing | ctDNA can detect minimal residual disease (MRD) and predict survival | [28] |
| 95 HCC; 45 cirrhotic | Surgery | TERT promoter mutation (C228T) | Droplet digital PCR (ddPCR) | Detection of mutated TERT promoter was associated with lower survival | [34] |
| 59 HCC | Surgery TACE RFA Systemic chemotherapy BSC | Single nucleotide variant (SNV) in a panel of 69 genes | Targeted-sequencing | Mutated MLH1 in plasma was associated with lower survival | [35] |
| 130 HCC | TACE Systemic chemotherapy | TERT promoter mutation | Droplet digital PCR (ddPCR) | Detection of mutated TERT promoter was associated with lower survival | [36] |
| 895 HCC | Surgery/NA | TP53 mutation (R249S) | Droplet digital PCR (ddPCR) | Detection of mutated TP53 was associated with lower survival | [37] |
| 22 HCC | TKI (tyrosine kinase inhibitors) | Genes of the PI3K/MTOR pathway | Targeted-sequencing and ddPCR | Mutations of genes in the PI3K/MTOR pathway are associated with lower survival in patients treated with TKI | [38] |
| Methylation Changes | |||||
| 72 HCC; 37 benign liver diseases; 41 healthy controls | - | APC, GSTP1, RASSF1A, and SFRP1 | Methylation-specific PCR | Methylation of RASSF1A was associated with poor survival | [39] |
| 1098 HCC; 835 controls | NA | 8-marker panel | Targeted bisulfite sequencing | Methylation-based classifier predicted survival | [40] |
| 10 HCC | TACE RFA Surgery | Methylation of GSTP1 and RASSF1A or TP53 mutation | Methylation-specific PCR and sanger sequencing | Detecting ctDNA in urine was feasible and predicted recurrence | [32] |
| 203 HCC; 104 chronic viral hepatitis B or C; 50 healthy controls | NA | APC, COX2, RASSF1A (+miR-203) | Methylation-specific PCR | Classifier predicted survival | [41] |
| 172 HCC | NA | LINE-1 | Methylation-specific PCR | Hypomethylation of LINE-1 was associated with lower survival | [42] |
| 155 HCC; 60 chronic HBV; 20 healthy controls | Surgery | IGFBP7 | Methylation-specific PCR | Methylation of IGFBP7 was associated with lower survival | [43] |
| 43 HCC (+347 HCC from TCGA Atlas); 5 cirrhotic; 6 benign liver lesions | - | CTCFL | Methylation-specific PCR | Hypomethylation of CTCFL was associated with higher recurrence and lower survival | [44] |
| Number of Patients | Treatment | Biomarkers | Technique | Main Finding | [Ref.] |
|---|---|---|---|---|---|
| miRNA | |||||
| 195 HCC 54 cirrhotic | Surgery Transplant TACE RFA sorafenib | miR-1 and miR-12 | qRT-PCR | Low level of miR-1 was associated with lower survival | [50] |
| 122 HCC | Surgery | miR-122 | qRT-PCR | Low level of miR-122 was associated with lower survival | [51] |
| 120 HCC | Surgery RFA | MiR-21, miR-26a, and miR-29a | qRT-PCR | Low levels of miR-26a and miR-29a were associated with lower survival | [52] |
| 30 HCC; 30 controls | Surgery | miR-155, miR-96 and miR-99a | qRT-PCR | High levels of miR-155 and miR-96 were associated with lower survival | [53] |
| 116 HCC | NA | Circulating miR | Whole miRNome profling | Low levels of miR-424-5p, miR-101-3p or high levels of miR-128, miR-139-5p, miR-382-5p and miR410 were associated with lower survival | [54] |
| 41 HCC; 20 controls | Surgery transplant | miR193a-5p | qRT-PCR | High level of miR193a-5p was associated with lower survival | [55] |
| 70 HBV-related HCC 70 HBV 50 healthy controls | Surgery | miRNA-223-3p | qRT-PCR | Low level of miRNA-223-3p was associated with lower survival | [56] |
| mRNA | |||||
| 50 HCC; 50 controls | Surgery | VEGF-165 | qRT-PCR | Detection of circulating VEGF mRNA (isoform 165) was associated with higher recurrence and recurrence-related mortality | [57] |
| 38 HCC | Surgery | AFP | qRT-PCR | Detection of AFP mRNA was associated with extrahepatic recurrence and shorter disease-free survival | [58] |
| 343 HCC | Surgery TACE RFA Systemic chemotherapy Radiotherapy BSC | AFP and hTERT | qRT-PCR | Detection of AFP mRNA or hTERT mRNA was not associated with survival | [59] |
| Exosomes | |||||
| 59 HCC | Transplant | miR-718 | qRT-PCR | Recurrence was associated with higher level of exosomal miR-718 | [60] |
| 30 HCC | Surgery | miR-665 | qRT-PCR | High level of exosomal miR-665 was associated with lower survival | [61] |
| 79 HCC | Surgery Transplant TACE RFA Sorafenib BSC | miR-21 and lncRNA-ATB | qRT-PCR | High levels of exosomal miR-21 and lncRNA-ATB were associated with lower survival | [62] |
| 126 HCC; 21 healthy controls | Surgery | miR-638 | qRT-PCR | Low level of exosomal miR-638 was associated with lower survival | [63] |
| 31 HCC; 3 CLD; 11 healthy controls | NA | RN7SL1 S fragment | qRT-PCR | High expression of RN7SL1 S fragment was associated with lower survival | [64] |
| 124 HCC; 100 healthy controls | Surgery | AKT3 | qRT-PCR | High level of exosomal circulating AKT3 was associated with higher recurrence and lower survival rates | [65] |
| 104 HCC; 55 CLD; 50 healthy controls | Surgery | miR-320a | qRT-PCR | Low serum exosomal miR-320a was associated with lower survival | [66] |
| Number of Patients | Treatment | Technique of Detection | Main Finding | [Ref.] |
|---|---|---|---|---|
| 44 HCC 30 HCV 39 cirrhosis 38 healthy controls | Surgery NA | Isolation by size of epithelial tumor cells (ISET) | Presence and number of detected CTCs were associated with shorter survival | [77] |
| 85 HCC 37 benign liver diseases 20 healthy volunteers 14 miscellaneous advanced cancers other than HCC | Surgery NA | Antibody-coated magnetic beads | Presence and number of detected CTCs correlated with tumor size, portal vein tumor thrombus, differentiation status, TNM stage and Milan criteria | [78] |
| 82 HCC | Surgery | Multicolor flow cytometry | Circulating cancer stem cells (CSC) are associated with higher rates of intra- and extra-hepatic recurrence, decreased recurrence-free survival (RFS) and overall survival (OS) rates | [79] |
| 96 HCC 31 healthy controls 21 viral hepatitis 8 cirrhosis | Surgery | Magnetic cell sorting (Lin28B) | Detection of CTCs expressing Lin28B was associated with early recurrence | [80] |
| 60 HCC | Surgery NA | Flow cytometry (ICAM-1) | Detection of CTCs expressing ICAM-1 was associated with shorter disease-free survival | [81] |
| 123 HCC | Surgery | EpCAM antibody-coated magnetic beads (CellSearch) | Detection of CTCs (EpCAM+) was associated with higher recurrence | [75] |
| 59 HCC 19 controls | NA | EpCAM antibody-coated magnetic beads (CellSearch) | Detection of CTCs was associated with lower overall survival | [82] |
| 122 HCC 120 controls | Surgery TACE Radiotherapy | EpCAM antibody-coated magnetic beads (CellSearch) | Peri-treatment decrease of detected CTC reflected treatment response | [83] |
| 109 HCC | Surgery TACE RFA sorafenib | Flow cytometry (ASGPR and CPS1) | pERK+/pAkt-CTCs correlated with progression-free survival and predicted response to systemic therapy (sorafenib) | [84] |
| 72 HCC | Surgery | EpCAM antibody-coated magnetic nanoparticals (MagVigen, Nvigen) | Detection of CTCs expressing AFP was associated with metastatic disease | [85] |
| 69 HCC 31 controls | Surgery Transplant TACE RFA Sorafenib BSC | Imaging flow cytometry (EpCAM, AFP, glypican-3 and DNA-PK together with analysis of size, morphology and DNA content) (ImageStream) | Detection of CTCs was associated with lower survival | [86] |
| 57 HCC | Surgery | EpCAM antibody-coated magnetic beads (CellSearch) | CTCs detection was associated with higher recurrence and lower recurrence-free survival after liver resection | [87] |
| 14 HCC 16 CCA 4 GBC | Surgery | SE-iFISH | Detection of small CTCs with CNV (chromosome 8) was associated with lower survival | [88] |
| 199 HCC | Surgery | Fluorescence-activated cell sorting (FACS) | Anterior approach was associated with a decreased dissemination of CTCs compared to conventional approach, resulting in poorer outcomes. | [89] |
| 73 HCC | Surgery | EpCAM antibody-coated magnetic beads (CellSearch) | Analyzes of blood samples collected in different vessels revealed a spatial heterogeneity of CTCs distribution whose biology was associated with recurrence pattern. | [90] |
| 130 HCC | Surgery TACE | qRT-PCR test platform | CTCs detection was associated with recurrence after liver resection | [91] |
| 112 HCC | Surgery | CanPatrolTM system (filtration by size) and Tri-color RNA-ISH assay | The presence of CTCs and the proportion of mesenchymal-CTC (M-CTCs) were associated with recurrence | [92] |
| 61 HCC 19 non-HCC | TACE TARE RFA Systemic therapy | Antibody-based platform | Vimentin (VIM)-positive CTCs predicted OS and faster recurrence after curative-intent surgical or locoregional therapy in potentially curable early-stage HCC | [93] |
| 139 HCC 23 controls | Surgery | EpCAM antibody-coated magnetic beads (CellSearch) | Surgical resection induces a release of CTCs | [94] |
| 105 HCC | Surgery | ISET | ΔCTCs is an independent predictor of lower survival and higher recurrence in patients | [95] |
| 85 HCC 27 non-HCC | Surgery | Flow cytometry (GPC3) | GPC3 positive-CTCs detection was associated with lower survival | [96] |
| 50 HCC | Transplant | Negative enrichment and immunofluorescence in situ hybridization (imFISH) | CTCs detection was associated with early recurrence after liver transplant | [97] |
| 137 HCC | Surgery | ISET | CTCs detection was associated with early recurrence after liver resection | [98] |
| 87 HCC 7 cirrhosis 8 healthy controls | Transplant Surgery TACE TARE RFA Systemic therapy | Antibody-based platform | Detection of CTCs expressing PD-L1 were associated with shorter OS and predicted response to immunotherapy | [99] |
| 128 HCC | Surgery ± TACE | EpCAM antibody-coated magnetic beads (CellSearch) | Adjuvant TACE provided survival and recurrence benefits in patients with positive preoperative CTCs | [100] |
| 193 HCC | Transplant | Antibody-based platform (ChimeraX®-i120) | CTCs detection was associated with recurrence after liver transplant | [101] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Labgaa, I.; Villanueva, A.; Dormond, O.; Demartines, N.; Melloul, E. The Role of Liquid Biopsy in Hepatocellular Carcinoma Prognostication. Cancers 2021, 13, 659. https://doi.org/10.3390/cancers13040659
Labgaa I, Villanueva A, Dormond O, Demartines N, Melloul E. The Role of Liquid Biopsy in Hepatocellular Carcinoma Prognostication. Cancers. 2021; 13(4):659. https://doi.org/10.3390/cancers13040659
Chicago/Turabian StyleLabgaa, Ismail, Augusto Villanueva, Olivier Dormond, Nicolas Demartines, and Emmanuel Melloul. 2021. "The Role of Liquid Biopsy in Hepatocellular Carcinoma Prognostication" Cancers 13, no. 4: 659. https://doi.org/10.3390/cancers13040659
APA StyleLabgaa, I., Villanueva, A., Dormond, O., Demartines, N., & Melloul, E. (2021). The Role of Liquid Biopsy in Hepatocellular Carcinoma Prognostication. Cancers, 13(4), 659. https://doi.org/10.3390/cancers13040659