Tracking the Progression of Triple Negative Mammary Tumors over Time by Chemometric Analysis of Urinary Volatile Organic Compounds
Abstract
:Simple Summary
Abstract
1. Introduction
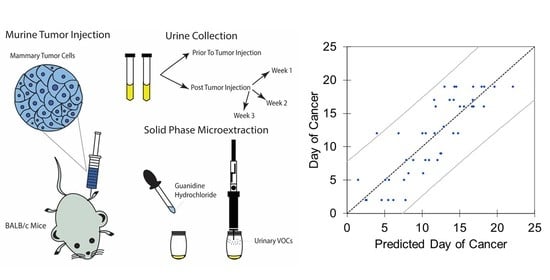
2. Materials and Methods
2.1. Materials and Instrumentation
2.2. Tumor Injection and Urine Collection
2.3. HS-SPME and GC-MS QTOF Analysis
2.4. Data Treatment and Chemometric Analyses
2.5. Regression Analyses
2.6. VOC Identification and Metabolic Pathway Analysis
3. Results
3.1. Urine Collection, Spectral Alignment and Data Normalization
3.2. Univariate Statistical Analysis
3.3. Multivariate Classification Analyses
3.4. Linear and Principal Component Regression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Kennecke, H.; Yerushalmi, R.; Woods, R.; Cheang, M.C.U.; Voduc, D.; Speers, C.H.; Nielsen, T.O.; Gelmon, K. Metastatic Behavior of Breast Cancer Subtypes. JCO 2010, 28, 3271–3277. [Google Scholar] [CrossRef]
- Diagnostic & Monitoring Tests|Cancer Testing|UNM Cancer Center. Available online: http://cancer.unm.edu/cancer/cancer-info/testing-overview/diagnostics-monitoring-tests/ (accessed on 11 May 2019).
- Radhakrishna, S.; Agarwal, S.; Parikh, P.M.; Kaur, K.; Panwar, S.; Sharma, S.; Dey, A.; Saxena, K.K.; Chandra, M.; Sud, S. Role of Magnetic Resonance Imaging in Breast Cancer Management. South Asian J. Cancer 2018, 7, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Thoeny, H.C.; Ross, B.D. Predicting and Monitoring Cancer Treatment Response with Diffusion-Weighted MRI. J. Magn. Reson. Imaging 2010, 32, 2–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taverna, G.; Tidu, L.; Grizzi, F.; Torri, V.; Mandressi, A.; Sardella, P.; La Torre, G.; Cocciolone, G.; Seveso, M.; Giusti, G.; et al. Olfactory System of Highly Trained Dogs Detects Prostate Cancer in Urine Samples. J. Urol. 2015, 193, 1382–1387. [Google Scholar] [CrossRef]
- Hackner, K.; Errhalt, P.; Mueller, M.R.; Speiser, M.; Marzluf, B.A.; Schulheim, A.; Schenk, P.; Bilek, J.; Doll, T. Canine Scent Detection for the Diagnosis of Lung Cancer in a Screening-like Situation. J. Breath Res. 2016, 10, 046003. [Google Scholar] [CrossRef]
- McCulloch, M.; Jezierski, T.; Broffman, M.; Hubbard, A.; Turner, K.; Janecki, T. Diagnostic Accuracy of Canine Scent Detection in Early- and Late-Stage Lung and Breast Cancers. Integr. Cancer 2006, 5, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R.T.; Schatz, C.B.; Myers, L.J.; Kosty, M.; Gonczy, C.; Kroener, J.; Tran, M.; Kurtzhals, P.; Heath, S.; Koziol, J.A.; et al. The Use of Canines in the Detection of Human Cancers. J. Altern. Complement Med. 2008, 14, 61–67. [Google Scholar] [CrossRef]
- Human Ovarian Carcinomas Detected by Specific Odor—György Horvath, Gunvor Af Klinteberg Järverud, Sven Järverud, István Horváth. 2008. Available online: https://journals.sagepub.com/doi/abs/10.1177/1534735408319058 (accessed on 8 June 2020).
- Jezierski, T.; Walczak, M.; Ligor, T.; Rudnicka, J.; Buszewski, B. Study of the Art: Canine Olfaction Used for Cancer Detection on the Basis of Breath Odour. Perspectives and Limitations. J. Breath Res. 2015, 9, 027001. [Google Scholar] [CrossRef]
- Jenkins, E.K.; DeChant, M.T.; Perry, E.B. When the Nose Doesn’t Know: Canine Olfactory Function Associated With Health, Management, and Potential Links to Microbiota. Front. Vet. Sci. 2018, 5. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janfaza, S.; Khorsand, B.; Nikkhah, M.; Zahiri, J. Digging Deeper into Volatile Organic Compounds Associated with Cancer. Biol. Methods Protoc. 2019, 4. [Google Scholar] [CrossRef]
- Saalberg, Y.; Wolff, M. VOC Breath Biomarkers in Lung Cancer. Clin. Chim. Acta 2016, 459, 5–9. [Google Scholar] [CrossRef]
- Khalid, T.; Aggio, R.; White, P.; De Lacy Costello, B.; Persad, R.; Al-Kateb, H.; Jones, P.; Probert, C.S.; Ratcliffe, N. Urinary Volatile Organic Compounds for the Detection of Prostate Cancer. PLoS ONE 2015, 10, e0143283. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Su, X.; Annabi, M.H.; Schreiter, B.R.; Prince, T.; Ackerman, A.; Morgas, S.; Mata, V.; Williams, H.; Lee, W.-Y. Application of Urinary Volatile Organic Compounds (VOCs) for the Diagnosis of Prostate Cancer. Clin. Genitourin. Cancer 2019, 17, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.R.; Pinto, J.; Azevedo, A.I.; Barros-Silva, D.; Jerónimo, C.; Henrique, R.; de Lourdes Bastos, M.; Guedes de Pinho, P.; Carvalho, M. Identification of a Biomarker Panel for Improvement of Prostate Cancer Diagnosis by Volatile Metabolic Profiling of Urine. Br. J. Cancer 2019, 121, 857–868. [Google Scholar] [CrossRef]
- Lima, A.R.; Araújo, A.M.; Pinto, J.; Jerónimo, C.; Henrique, R.; de Bastos, M.L.; Carvalho, M.; de Pinho, P.G. Discrimination between the Human Prostate Normal and Cancer Cell Exometabolome by GC-MS. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Phillips, M.; Cataneo, R.N.; Saunders, C.; Hope, P.; Schmitt, P.; Wai, J. Volatile Biomarkers in the Breath of Women with Breast Cancer. J. Breath Res. 2010, 4, 026003. [Google Scholar] [CrossRef]
- Peng, G.; Hakim, M.; Broza, Y.Y.; Billan, S.; Abdah-Bortnyak, R.; Kuten, A.; Tisch, U.; Haick, H. Detection of Lung, Breast, Colorectal, and Prostate Cancers from Exhaled Breath Using a Single Array of Nanosensors. Br. J. Cancer 2010, 103, 542–551. [Google Scholar] [CrossRef]
- Lima, A.R.; Pinto, J.; Carvalho-Maia, C.; Jerónimo, C.; Henrique, R.; de Bastos, M.L.; Carvalho, M.; Guedes de Pinho, P. A Panel of Urinary Volatile Biomarkers for Differential Diagnosis of Prostate Cancer from Other Urological Cancers. Cancers 2020, 12, 2017. [Google Scholar] [CrossRef]
- Beretov, J.; Wasinger, V.C.; Millar, E.K.A.; Schwartz, P.; Graham, P.H.; Li, Y. Proteomic Analysis of Urine to Identify Breast Cancer Biomarker Candidates Using a Label-Free LC-MS/MS Approach. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Cala, M.; Aldana, J.; Sánchez, J.; Guio, J.; Meesters, R.J.W. Urinary Metabolite and Lipid Alterations in Colombian Hispanic Women with Breast Cancer: A Pilot Study. J. Pharm. Biomed. Anal. 2018, 152, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Hirschfeld, M.; Rücker, G.; Weiß, D.; Berner, K.; Ritter, A.; Jäger, M.; Erbes, T. Urinary Exosomal MicroRNAs as Potential Non-Invasive Biomarkers in Breast Cancer Detection. Mol. Diagn. 2020, 24, 215–232. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.L.; Passos, M.; Câmara, J.S. Solid Phase Microextraction, Mass Spectrometry and Metabolomic Approaches for Detection of Potential Urinary Cancer Biomarkers—A Powerful Strategy for Breast Cancer Diagnosis. Talanta 2012, 89, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.L.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Implementing a Central Composite Design for the Optimization of Solid Phase Microextraction to Establish the Urinary Volatomic Expression: A First Approach for Breast Cancer. Metabolomics 2019, 15, 64. [Google Scholar] [CrossRef] [PubMed]
- Woollam, M.; Teli, M.; Angarita-Rivera, P.; Liu, S.; Siegel, A.P.; Yokota, H.; Agarwal, M. Detection of Volatile Organic Compounds (VOCs) in Urine via Gas Chromatography-Mass Spectrometry QTOF to Differentiate Between Localized and Metastatic Models of Breast Cancer. Sci. Rep. 2019, 9, 2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woollam, M.; Teli, M.; Liu, S.; Daneshkhah, A.; Siegel, A.P.; Yokota, H.; Agarwal, M. Urinary Volatile Terpenes Analyzed by Gas Chromatography–Mass Spectrometry to Monitor Breast Cancer Treatment Efficacy in Mice. J. Proteome Res. 2020, 19, 1913–1922. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Chen, A.; Teli, M.; Kondo, R.; Jalali, A.; Fan, Y.; Liu, S.; Zhao, X.; Siegel, A.; et al. Pitavastatin Slows Tumor Progression and Alters Urine-Derived Volatile Organic Compounds through the Mevalonate Pathway. FASEB J. 2019, 33, 13710–13721. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Jalali, A.; Chen, A.; Zhao, X.; Liu, S.; Teli, M.; Guo, Y.; Li, F.; Li, J.; Siegel, A.; et al. Skeletal Loading Regulates Breast Cancer-Associated Osteolysis in a Loading Intensity-Dependent Fashion. Bone Res. 2020, 8, 9. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, T.; Ricci, M.; Scarpi, E.; Bongiovanni, A.; Ricci, R.; Riva, N.; Liverani, C.; De Vita, A.; La Manna, F.; Oboldi, D.; et al. RANKL: A Promising Circulating Marker for Bone Metastasis Response. Oncol. Lett. 2016, 12, 2970–2975. [Google Scholar] [CrossRef] [Green Version]
- Tulotta, C.; Groenewoud, A.; Snaar-Jagalska, B.E.; Ottewell, P. Animal Models of Breast Cancer Bone Metastasis. Bone Res. Protoc. 2019, 309–330. [Google Scholar] [CrossRef]
- Liu, S.; Fan, Y.; Chen, A.; Jalali, A.; Minami, K.; Ogawa, K.; Nakshatri, H.; Li, B.-Y.; Yokota, H. Osteocyte-Driven Downregulation of Snail Restrains Effects of Drd2 Inhibitors on Mammary Tumor Cells. Cancer Res. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwak, J.; Grigsby, C.C.; Rizki, M.M.; Preti, G.; Köksal, M.; Josue, J.; Yamazaki, K.; Beauchamp, G.K. Differential Binding between Volatile Ligands and Major Urinary Proteins Due to Genetic Variation in Mice. Physiol. Behav. 2012, 107, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Tang, J.; Yang, Q.; Li, S.; Cui, X.; Li, Y.; Chen, Y.; Xue, W.; Li, X.; Zhu, F. NOREVA: Normalization and Evaluation of MS-Based Metabolomics Data. Nucleic Acids Res. 2017, 45, W162–W170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Siegel, A.P.; Daneshkhah, A.; Hardin, D.S.; Shrestha, S.; Varahramyan, K.; Agarwal, M. Analyzing Breath Samples of Hypoglycemic Events in Type 1 Diabetes Patients: Towards Developing an Alternative to Diabetes Alert Dogs. J. Breath Res. 2017, 11, 026007. [Google Scholar] [CrossRef] [PubMed]
- Babyak, M.A. What You See May Not Be What You Get: A Brief, Nontechnical Introduction to Overfitting in Regression-Type Models. Psychosom. Med. 2004, 66, 411–421. [Google Scholar] [PubMed] [Green Version]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Qiu, Y.-Q. KEGG Pathway Database. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., Yokota, H., Eds.; Springer: New York, NY, USA, 2013; pp. 1068–1069. ISBN 978-1-4419-9863-7. [Google Scholar]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational Biomarker Discovery in Clinical Metabolomics: An Introductory Tutorial. Metabolomics 2013, 9, 280–299. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.L.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Volatile Metabolomic Signature of Human Breast Cancer Cell Lines. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Yang, D.; Brock, G.; Knipp, R.J.; Bousamra, M.; Nantz, M.H.; Fu, X.-A. Breath Carbonyl Compounds as Biomarkers of Lung Cancer. Lung Cancer 2015, 90, 92–97. [Google Scholar] [CrossRef]
- Santos, P.M.; del Nogal Sánchez, M.; Pozas, Á.P.C.; Pavón, J.L.P.; Cordero, B.M. Determination of Ketones and Ethyl Acetate—a Preliminary Study for the Discrimination of Patients with Lung Cancer. Anal. Bioanal. Chem. 2017, 409, 5689–5696. [Google Scholar] [CrossRef]
- Minh, T.D.C.; Blake, D.R.; Galassetti, P.R. The Clinical Potential of Exhaled Breath Analysis For Diabetes Mellitus. Diabetes Res. Clin. Pr. 2012, 97, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Fan, Y.; Liu, S.; Woollam, M.D.; Sun, X.; Murao, E.; Zha, R.; Prakash, R.; Park, C.; Siegel, A.P.; et al. Loading-Induced Antitumor Capability of Murine and Human Urine. FASEB J. 2020. [Google Scholar] [CrossRef] [Green Version]
- Clendening, J.W.; Pandyra, A.; Boutros, P.C.; Ghamrasni, S.E.; Khosravi, F.; Trentin, G.A.; Martirosyan, A.; Hakem, A.; Hakem, R.; Jurisica, I.; et al. Dysregulation of the Mevalonate Pathway Promotes Transformation. PNAS 2010, 107, 15051–15056. [Google Scholar] [CrossRef] [Green Version]
- Pampalakis, G.; Obasuyi, O.; Papadodima, O.; Chatziioannou, A.; Zoumpourlis, V.; Sotiropoulou, G. The KLK5 Protease Suppresses Breast Cancer by Repressing the Mevalonate Pathway. Oncotarget 2013, 5, 2390–2403. [Google Scholar] [CrossRef] [Green Version]
- Ahn, K.S.; Sethi, G.; Chaturvedi, M.M.; Aggarwal, B.B. Simvastatin, 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase Inhibitor, Suppresses Osteoclastogenesis Induced by Receptor Activator of Nuclear Factor-ΚB Ligand through Modulation of NF-ΚB Pathway. Int. J. Cancer 2008, 123, 1733–1740. [Google Scholar] [CrossRef]
- Mandal, C.C.; Ghosh-Choudhury, N.; Yoneda, T.; Choudhury, G.G.; Ghosh-Choudhury, N. Simvastatin Prevents Skeletal Metastasis of Breast Cancer by an Antagonistic Interplay between P53 and CD44. J. Biol. Chem. 2011, 286, 11314–11327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llaverias, G.; Danilo, C.; Mercier, I.; Daumer, K.; Capozza, F.; Williams, T.M.; Sotgia, F.; Lisanti, M.P.; Frank, P.G. Role of Cholesterol in the Development and Progression of Breast Cancer. Am. J. Pathol. 2011, 178, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Gould, M.N. Prevention and Therapy of Mammary Cancer by Monoterpenes. J. Cell. Biochem. 1995, 59, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Paduch, R.; Kandefer-Szerszeń, M.; Trytek, M.; Fiedurek, J. Terpenes: Substances Useful in Human Healthcare. Arch. Immunol. Ther. Exp. 2007, 55, 315. [Google Scholar] [CrossRef] [PubMed]
- Neighbors, J.D. The Mevalonate Pathway and Terpenes: A Diversity of Chemopreventatives. Curr. Pharm. Rep. 2018, 4, 157–169. [Google Scholar] [CrossRef]
- Jafari, S.; Saeidnia, S.; Abdollahi, M. Role of Natural Phenolic Compounds in Cancer Chemoprevention via Regulation of the Cell Cycle. Curr. Pharm. Biotechnol. 2014, 15, 409–421. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woollam, M.; Wang, L.; Grocki, P.; Liu, S.; Siegel, A.P.; Kalra, M.; Goodpaster, J.V.; Yokota, H.; Agarwal, M. Tracking the Progression of Triple Negative Mammary Tumors over Time by Chemometric Analysis of Urinary Volatile Organic Compounds. Cancers 2021, 13, 1462. https://doi.org/10.3390/cancers13061462
Woollam M, Wang L, Grocki P, Liu S, Siegel AP, Kalra M, Goodpaster JV, Yokota H, Agarwal M. Tracking the Progression of Triple Negative Mammary Tumors over Time by Chemometric Analysis of Urinary Volatile Organic Compounds. Cancers. 2021; 13(6):1462. https://doi.org/10.3390/cancers13061462
Chicago/Turabian StyleWoollam, Mark, Luqi Wang, Paul Grocki, Shengzhi Liu, Amanda P. Siegel, Maitri Kalra, John V. Goodpaster, Hiroki Yokota, and Mangilal Agarwal. 2021. "Tracking the Progression of Triple Negative Mammary Tumors over Time by Chemometric Analysis of Urinary Volatile Organic Compounds" Cancers 13, no. 6: 1462. https://doi.org/10.3390/cancers13061462
APA StyleWoollam, M., Wang, L., Grocki, P., Liu, S., Siegel, A. P., Kalra, M., Goodpaster, J. V., Yokota, H., & Agarwal, M. (2021). Tracking the Progression of Triple Negative Mammary Tumors over Time by Chemometric Analysis of Urinary Volatile Organic Compounds. Cancers, 13(6), 1462. https://doi.org/10.3390/cancers13061462