Tyrosine Kinase c-MET as Therapeutic Target for Radiosensitization of Head and Neck Squamous Cell Carcinomas
Abstract
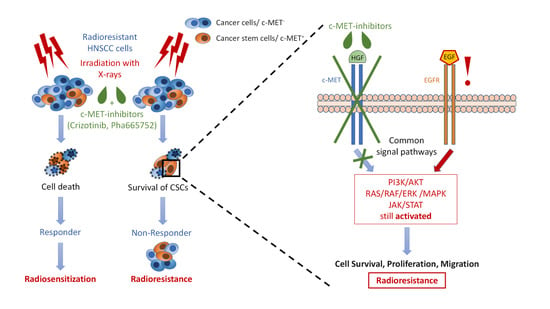
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Colony Formation Assay
2.3. Cell Irradiation
2.4. Viability Assay
2.5. Western Blot Analysis
2.6. Flow Cytometry Analysis and Fluorescence-Activated Cell Sorting (FACS)
2.7. Immunofluorescence Microscopy
2.8. siRNA-Mediated Gene Knock-Down
2.9. Sphere-Formation Assay
2.10. Xenograft Tumor Growth
2.11. Kinome Profiling
2.12. In Silico Gene Expression and Proteome Analysis
2.13. Statistics
3. Results
3.1. High c-MET Expression Characterizes a Radioresistant Subpopulation in HNSCC
3.2. c-MET-Expressing HNSCC Cells Are Less Sensitive to Irradiation without Affecting DNA Repair
3.3. The c-MET-Expressing Population in HNSCC Is Dynamically Regulated upon Irradiation and Is Characterized by Stem-Like Features
3.4. Compensatory Mechanisms Overcoming c-MET-Mediated Radioresponse Uncovered by MET-Specific Gene Knock-Down
3.5. Chemical c-MET Targeting for HNSCC Radiosensitization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rettig, E.M.; D’Souza, G. Epidemiology of Head and Neck Cancer. Surg. Oncol. Clin. N. Am. 2015, 24, 379–396. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021. [Google Scholar] [CrossRef] [PubMed]
- Gatta, G.; Botta, L.; Sánchez, M.J.; Anderson, L.A.; Pierannunzio, D.; Licitra, L.; EUROCARE Working Group. Prognoses and Improvement for Head and Neck Cancers Diagnosed in Europe in Early 2000s: The EUROCARE-5 Population-Based Study. Eur. J. Cancer 2015, 51, 2130–2143. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Puri, D.R.; Blanco, A.I.; Chao, K.S.C. Intensity-Modulated Radiation Therapy in Head and Neck Cancers: An Update. Head Neck 2007, 29, 387–400. [Google Scholar] [CrossRef]
- Argiris, A.; Li, S.; Savvides, P.; Ohr, J.P.; Gilbert, J.; Levine, M.A.; Chakravarti, A.; Haigentz, M., Jr.; Saba, N.F.; Ikpeazu, C.V.; et al. Phase III Randomized Trial of Chemotherapy with or without Bevacizumab in Patients with Recurrent or Metastatic Head and Neck Cancer. J. Clin. Oncol. 2019, 37, 3266–3274. [Google Scholar] [CrossRef]
- Muzaffar, J.; Bari, S.; Kirtane, K.; Chung, C.H. Recent Advances and Future Directions in Clinical Management of Head and Neck Squamous Cell Carcinoma. Cancers 2021, 13, 338. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Sougnez, C.; Lichtenstein, L.; Cibulskis, K.; Lander, E.; Gabriel, S.B.; Getz, G.; Ally, A.; Balasundaram, M.; Birol, I.; et al. Comprehensive Genomic Characterization of Head and Neck Squamous Cell Carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [Green Version]
- Chung, C.H.; Guthrie, V.B.; Masica, D.L.; Tokheim, C.; Kang, H.; Richmon, J.; Agrawal, N.; Fakhry, C.; Quon, H.; Subramaniam, R.M.; et al. Genomic Alterations in Head and Neck Squamous Cell Carcinoma Determined by Cancer Gene-Targeted Sequencing. Ann. Oncol. 2015, 26, 1216–1223. [Google Scholar] [CrossRef]
- Kalyankrishna, S.; Grandis, J.R. Epidermal Growth Factor Receptor Biology in Head and Neck Cancer. J. Clin. Oncol. 2006, 24, 2666–2672. [Google Scholar] [CrossRef] [PubMed]
- Ganci, F.; Sacconi, A.; Manciocco, V.; Spriano, G.; Fontemaggi, G.; Carlini, P.; Blandino, G. Radioresistance in Head and Neck Squamous Cell Carcinoma—Possible Molecular Markers for Local Recurrence and New Putative Therapeutic Strategies. Contemp. Issues Head Neck Cancer Manag. 2015, 3–34. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Fisher, G.J. Role of Met Axis in Head and Neck Cancer. Cancers 2013, 5, 1601. [Google Scholar] [CrossRef] [Green Version]
- Alsahafi, E.; Begg, K.; Amelio, I.; Raulf, N.; Lucarelli, P.; Sauter, T.; Tavassoli, M. Clinical Update on Head and Neck Cancer: Molecular Biology and Ongoing Challenges. Cell Death Dis. 2019, 10, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linge, A.; Löck, S.; Gudziol, V.; Nowak, A.; Lohaus, F.; von Neubeck, C.; Jütz, M.; Abdollahi, A.; Debus, J.; Tinhofer, I.; et al. Low Cancer Stem Cell Marker Expression and Low Hypoxia Identify Good Prognosis Subgroups in HPV(-) HNSCC after Postoperative Radiochemotherapy: A Multicenter Study of the DKTK-ROG. Clin. Cancer Res. 2016, 22, 2639–2649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothenberger, N.J.; Stabile, L.P. Hepatocyte Growth Factor/c-Met Signaling in Head and Neck Cancer and Implications for Treatment. Cancers 2017, 9, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Comoglio, P.M.; Giordano, S.; Trusolino, L. Drug Development of MET Inhibitors: Targeting Oncogene Addiction and Expedience. Nat. Rev. Drug Discov. 2008, 7, 504–516. [Google Scholar] [CrossRef]
- Toschi, L.; Jänne, P.A. Single-Agent and Combination Therapeutic Strategies to Inhibit Hepatocyte Growth Factor/MET Signaling in Cancer. Clin. Cancer Res. 2008, 14, 5941–5946. [Google Scholar] [CrossRef] [Green Version]
- Ponzetto, C.; Bardelli, A.; Zhen, Z.; Maina, F.; dalla Zonca, P.; Giordano, S.; Graziani, A.; Panayotou, G.; Comoglio, P.M. A Multifunctional Docking Site Mediates Signaling and Transformation by the Hepatocyte Growth Factor/Scatter Factor Receptor Family. Cell 1994, 77, 261–271. [Google Scholar] [CrossRef]
- Sachs, M.; Brohmann, H.; Zechner, D.; Müller, T.; Hülsken, J.; Walther, I.; Schaeper, U.; Birchmeier, C.; Birchmeier, W. Essential Role of Gab1 for Signaling by the C-Met Receptor in Vivo. J. Cell Biol. 2000, 150, 1375–1384. [Google Scholar] [CrossRef] [Green Version]
- Baldanzi, G.; Graziani, A. Physiological Signaling and Structure of the HGF Receptor MET. Biomedicines 2014, 3, 1–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bladt, F.; Riethmacher, D.; Isenmann, S.; Aguzzi, A.; Birchmeier, C. Essential Role for the C- Met Receptor in the Migration of Myogenic Precursor Cells into the Limb Bud. Nature 1995, 376, 768–771. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Nishizawa, T.; Hagiya, M.; Seki, T.; Shimonishi, M.; Sugimura, A.; Tashiro, K.; Shimizu, S. Molecular Cloning and Expression of Human Hepatocyte Growth Factor. Nature 1989, 342, 440–443. [Google Scholar] [CrossRef] [PubMed]
- Bonner, J.; Giralt, J.; Harari, P.; Spencer, S.; Schulten, J.; Hossain, A.; Chang, S.-C.; Chin, S.; Baselga, J. Cetuximab and Radiotherapy in Laryngeal Preservation for Cancers of the Larynx and Hypopharynx: A Secondary Analysis of a Randomized Clinical Trial. JAMA Otolaryngol. Head Neck Surg. 2016, 142, 842–849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takes, R.P.; Baatenburg de Jong, R.J.; Schuuring, E.; Litvinov, S.V.; Hermans, J.; Van Krieken, J.H. Differences in Expression of Oncogenes and Tumor Suppressor Genes in Different Sites of Head and Neck Squamous Cell. Anticancer Res. 1998, 18, 4793–4800. [Google Scholar] [PubMed]
- Mehanna, H.; Robinson, M.; Hartley, A.; Kong, A.; Foran, B.; Fulton-Lieuw, T.; Dalby, M.; Mistry, P.; Sen, M.; O’Toole, L.; et al. Radiotherapy plus Cisplatin or Cetuximab in Low-Risk Human Papillomavirus-Positive Oropharyngeal Cancer (De-ESCALaTE HPV): An Open-Label Randomised Controlled Phase 3 Trial. Lancet 2019, 393, 51–60. [Google Scholar] [CrossRef] [Green Version]
- Gebre-Medhin, M.; Brun, E.; Engström, P.; Haugen Cange, H.; Hammarstedt-Nordenvall, L.; Reizenstein, J.; Nyman, J.; Abel, E.; Friesland, S.; Sjödin, H.; et al. ARTSCAN III: A Randomized Phase III Study Comparing Chemoradiotherapy With Cisplatin Versus Cetuximab in Patients With Locoregionally Advanced Head and Neck Squamous Cell Cancer. J. Clin. Oncol. 2021, 39, 38–47. [Google Scholar] [CrossRef]
- Montagut, C.; Dalmases, A.; Bellosillo, B.; Crespo, M.; Pairet, S.; Iglesias, M.; Salido, M.; Gallen, M.; Marsters, S.; Tsai, S.P.; et al. Identification of a Mutation in the Extracellular Domain of the Epidermal Growth Factor Receptor Conferring Cetuximab Resistance in Colorectal Cancer. Nat. Med. 2012, 18, 221–223. [Google Scholar] [CrossRef]
- Da Costa, A.A.B.A.; Costa, F.D.; Araújo, D.V.; Camandaroba, M.P.G.; de Jesus, V.H.F.; Oliveira, A.; Alves, A.C.F.; Stecca, C.; Machado, L.; de Oliveira, A.C.F.; et al. The Roles of PTEN, CMET, and P16 in Resistance to Cetuximab in Head and Neck Squamous Cell Carcinoma. Med. Oncol. 2018, 36, 8. [Google Scholar] [CrossRef]
- Scagliotti, G.V.; Novello, S.; von Pawel, J. The Emerging Role of MET/HGF Inhibitors in Oncology. Cancer Treat. Rev. 2013, 39, 793–801. [Google Scholar] [CrossRef]
- Huang, X.; Li, E.; Shen, H.; Wang, X.; Tang, T.; Zhang, X.; Xu, J.; Tang, Z.; Guo, C.; Bai, X.; et al. Targeting the HGF/MET Axis in Cancer Therapy: Challenges in Resistance and Opportunities for Improvement. Front. Cell Dev. Biol. 2020, 8, 152. [Google Scholar] [CrossRef]
- Puccini, A.; Marín-Ramos, N.I.; Bergamo, F.; Schirripa, M.; Lonardi, S.; Lenz, H.-J.; Loupakis, F.; Battaglin, F. Safety and Tolerability of C-MET Inhibitors in Cancer. Drug Saf. 2019, 42, 211–233. [Google Scholar] [CrossRef]
- Wilson, G.D.; Thibodeau, B.J.; Fortier, L.E.; Pruetz, B.L.; Galoforo, S.; Marples, B.; Baschnagel, A.M.; Akervall, J.; Huang, J. Cancer Stem Cell Signaling during Repopulation in Head and Neck Cancer. Available online: https://www.hindawi.com/journals/sci/2016/1894782/ (accessed on 7 January 2021).
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer Stem Cells and Radioresistance: DNA Repair and Beyond. Cancers 2019, 11, 862. [Google Scholar] [CrossRef] [Green Version]
- Digomann, D.; Kurth, I.; Tyutyunnykova, A.; Chen, O.; Löck, S.; Gorodetska, I.; Peitzsch, C.; Skvortsova, I.-I.; Negro, G.; Aschenbrenner, B.; et al. The CD98 Heavy Chain Is a Marker and Regulator of Head and Neck Squamous Cell Carcinoma Radiosensitivity. Clin. Cancer Res. 2019, 25, 3152–3163. [Google Scholar] [CrossRef] [PubMed]
- Linge, A.; Löck, S.; Krenn, C.; Appold, S.; Lohaus, F.; Nowak, A.; Gudziol, V.; Baretton, G.B.; Buchholz, F.; Baumann, M.; et al. Independent Validation of the Prognostic Value of Cancer Stem Cell Marker Expression and Hypoxia-Induced Gene Expression for Patients with Locally Advanced HNSCC after Postoperative Radiotherapy. Clin. Transl. Radiat. Oncol. 2016, 1, 19–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linge, A.; Lohaus, F.; Löck, S.; Nowak, A.; Gudziol, V.; Valentini, C.; von Neubeck, C.; Jütz, M.; Tinhofer, I.; Budach, V.; et al. HPV Status, Cancer Stem Cell Marker Expression, Hypoxia Gene Signatures and Tumour Volume Identify Good Prognosis Subgroups in Patients with HNSCC after Primary Radiochemotherapy: A Multicentre Retrospective Study of the German Cancer Consortium Radiation Oncology Group (DKTK-ROG). Radiother. Oncol. 2016, 121, 364–373. [Google Scholar] [CrossRef]
- Kurth, I.; Hein, L.; Mäbert, K.; Peitzsch, C.; Koi, L.; Cojoc, M.; Kunz-Schughart, L.; Baumann, M.; Dubrovska, A. Cancer Stem Cell Related Markers of Radioresistance in Head and Neck Squamous Cell Carcinoma. Oncotarget 2015, 6, 34494–34509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prince, M.E.P.; Ailles, L.E. Cancer Stem Cells in Head and Neck Squamous Cell Cancer. J. Clin. Oncol. 2008, 26, 2871–2875. [Google Scholar] [CrossRef]
- Zhang, Z.; Filho, M.S.; Nör, J.E. The Biology of Head and Neck Cancer Stem Cells. Oral. Oncol. 2012, 48, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Helbig, L.; Koi, L.; Brüchner, K.; Gurtner, K.; Hess-Stumpp, H.; Unterschemmann, K.; Baumann, M.; Zips, D.; Yaromina, A. BAY 87-2243, a Novel Inhibitor of Hypoxia-Induced Gene Activation, Improves Local Tumor Control after Fractionated Irradiation in a Schedule-Dependent Manner in Head and Neck Human Xenografts. Radiat. Oncol. 2014, 9, 207. [Google Scholar] [CrossRef] [Green Version]
- Koi, L.; Bergmann, R.; Brüchner, K.; Pietzsch, J.; Pietzsch, H.-J.; Krause, M.; Steinbach, J.; Zips, D.; Baumann, M. Radiolabeled Anti-EGFR-Antibody Improves Local Tumor Control after External Beam Radiotherapy and Offers Theragnostic Potential. Radiother. Oncol. 2014, 110, 362–369. [Google Scholar] [CrossRef]
- Koi, L.; Löck, S.; Linge, A.; Thurow, C.; Hering, S.; Baumann, M.; Krause, M.; Gurtner, K. EGFR-Amplification plus Gene Expression Profiling Predicts Response to Combined Radiotherapy with EGFR-Inhibition: A Preclinical Trial in 10 HNSCC-Tumour-Xenograft Models. Radiother. Oncol. 2017, 124, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, K.; Molfenter, B.; Laureano, N.K.; Tawk, B.; Bieg, M.; Hostench, X.P.; Weichenhan, D.; Ullrich, N.D.; Shang, V.; Richter, D.; et al. Somatic Mutations and Promotor Methylation of the Ryanodine Receptor 2 Is a Common Event in the Pathogenesis of Head and Neck Cancer. Int. J. Cancer 2019, 145, 3299–3310. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Shen, Y.; Hostench, X.P.; Bieg, M.; Plath, M.; Ishaque, N.; Eils, R.; Freier, K.; Weichert, W.; Zaoui, K.; et al. Integrative Analysis of Multi-Omics Data Identified EGFR and PTGS2 as Key Nodes in a Gene Regulatory Network Related to Immune Phenotypes in Head and Neck Cancer. Clin. Cancer Res. 2020, 26, 3616–3628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linge, A.; Schmidt, S.; Lohaus, F.; Krenn, C.; Bandurska-Luque, A.; Platzek, I.; von Neubeck, C.; Appold, S.; Nowak, A.; Gudziol, V.; et al. Independent Validation of Tumour Volume, Cancer Stem Cell Markers and Hypoxia-Associated Gene Expressions for HNSCC after Primary Radiochemotherapy. Clin. Transl. Radiat. Oncol. 2019, 16, 40–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaromina, A.; Krause, M.; Thames, H.; Rosner, A.; Krause, M.; Hessel, F.; Grenman, R.; Zips, D.; Baumann, M. Pre-Treatment Number of Clonogenic Cells and Their Radiosensitivity Are Major Determinants of Local Tumour Control after Fractionated Irradiation. Radiother. Oncol. 2007, 83, 304–310. [Google Scholar] [CrossRef]
- De Bacco, F.; Casanova, E.; Medico, E.; Pellegatta, S.; Orzan, F.; Albano, R.; Luraghi, P.; Reato, G.; D’Ambrosio, A.; Porrati, P.; et al. The MET Oncogene Is a Functional Marker of a Glioblastoma Stem Cell Subtype. Cancer Res. 2012, 72, 4537–4550. [Google Scholar] [CrossRef] [Green Version]
- Gastaldi, S.; Sassi, F.; Accornero, P.; Torti, D.; Galimi, F.; Migliardi, G.; Molyneux, G.; Perera, T.; Comoglio, P.M.; Boccaccio, C.; et al. Met Signaling Regulates Growth, Repopulating Potential and Basal Cell-Fate Commitment of Mammary Luminal Progenitors: Implications for Basal-like Breast Cancer. Oncogene 2013, 32, 1428–1440. [Google Scholar] [CrossRef] [Green Version]
- Saini, M.; Verma, A.; Mathew, S.J. SPRY2 Is a Novel MET Interactor That Regulates Metastatic Potential and Differentiation in Rhabdomyosarcoma. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Corso, S.; Migliore, C.; Ghiso, E.; De Rosa, G.; Comoglio, P.M.; Giordano, S. Silencing the MET Oncogene Leads to Regression of Experimental Tumors and Metastases. Oncogene 2008, 27, 684–693. [Google Scholar] [CrossRef] [Green Version]
- Peschard, P.; Park, M. From Tpr-Met to Met, Tumorigenesis and Tubes. Oncogene 2007, 26, 1276–1285. [Google Scholar] [CrossRef] [Green Version]
- Migliore, C.; Giordano, S. Molecular Cancer Therapy: Can Our Expectation Be MET? Eur. J. Cancer 2008, 44, 641–651. [Google Scholar] [CrossRef]
- Trusolino, L.; Bertotti, A.; Comoglio, P.M. MET Signalling: Principles and Functions in Development, Organ Regeneration and Cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Eder, J.P.; Vande Woude, G.F.; Boerner, S.A.; LoRusso, P.M. Novel Therapeutic Inhibitors of the C-Met Signaling Pathway in Cancer. Clin. Cancer Res. 2009, 15, 2207–2214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baschnagel, A.M.; Williams, L.; Hanna, A.; Chen, P.Y.; Krauss, D.J.; Pruetz, B.L.; Akervall, J.; Wilson, G.D. C-Met Expression Is a Marker of Poor Prognosis in Patients with Locally Advanced Head and Neck Squamous Cell Carcinoma Treated with Chemoradiation. Int. J. Radiat. Oncol. Biol. Phys. 2014, 88, 701–707. [Google Scholar] [CrossRef]
- Akervall, J.; Guo, X.; Qian, C.-N.; Schoumans, J.; Leeser, B.; Kort, E.; Cole, A.; Resau, J.; Bradford, C.; Carey, T.; et al. Genetic and Expression Profiles of Squamous Cell Carcinoma of the Head and Neck Correlate with Cisplatin Sensitivity and Resistance in Cell Lines and Patients. Clin. Cancer Res. 2004, 10, 8204–8213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madoz-Gúrpide, J.; Zazo, S.; Chamizo, C.; Casado, V.; Caramés, C.; Gavín, E.; Cristóbal, I.; García-Foncillas, J.; Rojo, F. Activation of MET Pathway Predicts Poor Outcome to Cetuximab in Patients with Recurrent or Metastatic Head and Neck Cancer. J. Transl. Med. 2015, 13, 282. [Google Scholar] [CrossRef]
- Wong, S.J.; Heron, D.E.; Stenson, K.; Ling, D.C.; Vargo, J.A. Locoregional Recurrent or Second Primary Head and Neck Cancer: Management Strategies and Challenges. Am. Soc. Clin. Oncol. Educ. Book 2016, 36, e284–e292. [Google Scholar] [CrossRef] [PubMed]
- Bensimon, A.; Koch, J.P.; Francica, P.; Roth, S.M.; Riedo, R.; Glück, A.A.; Orlando, E.; Blaukat, A.; Aebersold, D.M.; Zimmer, Y.; et al. Deciphering MET-Dependent Modulation of Global Cellular Responses to DNA Damage by Quantitative Phosphoproteomics. Mol. Oncol. 2020, 14, 1185–1206. [Google Scholar] [CrossRef]
- Seiwert, T.; Sarantopoulos, J.; Kallender, H.; McCallum, S.; Keer, H.N.; Blumenschein, G. Phase II Trial of Single-Agent Foretinib (GSK1363089) in Patients with Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck. Investig. New Drugs 2013, 31, 417–424. [Google Scholar] [CrossRef] [Green Version]
- Blumenschein, G.R.; Mills, G.B.; Gonzalez-Angulo, A.M. Targeting the Hepatocyte Growth Factor-CMET Axis in Cancer Therapy. J. Clin. Oncol. 2012, 30, 3287–3296. [Google Scholar] [CrossRef] [Green Version]
- Raghav, K.P.; Wang, W.; Liu, S.; Chavez-MacGregor, M.; Meng, X.; Hortobagyi, G.N.; Mills, G.B.; Meric-Bernstam, F.; Blumenschein, G.R.; Gonzalez-Angulo, A.M. CMET and Phospho-CMET Protein Levels in Breast Cancers and Survival Outcomes. Clin. Cancer Res. 2012, 18, 2269–2277. [Google Scholar] [CrossRef] [Green Version]
- Spigel, D.R.; Reynolds, C.; Waterhouse, D.; Garon, E.B.; Chandler, J.; Babu, S.; Thurmes, P.; Spira, A.; Jotte, R.; Zhu, J.; et al. Phase 1/2 Study of the Safety and Tolerability of Nivolumab Plus Crizotinib for the First-Line Treatment of Anaplastic Lymphoma Kinase Translocation—Positive Advanced Non-Small Cell Lung Cancer (CheckMate 370). J. Thorac. Oncol. 2018, 13, 682–688. [Google Scholar] [CrossRef] [Green Version]
- Gherardi, E.; Youles, M.E.; Miguel, R.N.; Blundell, T.L.; Iamele, L.; Gough, J.; Bandyopadhyay, A.; Hartmann, G.; Butler, P.J.G. Functional Map and Domain Structure of MET, the Product of the c-Met Protooncogene and Receptor for Hepatocyte Growth Factor/Scatter Factor. Proc. Natl. Acad. Sci. USA 2003, 100, 12039–12044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baschnagel, A.M.; Galoforo, S.; Thibodeau, B.J.; Ahmed, S.; Nirmal, S.; Akervall, J.; Wilson, G.D. Crizotinib Fails to Enhance the Effect of Radiation in Head and Neck Squamous Cell Carcinoma Xenografts. Anticancer Res. 2015, 35, 5973–5982. [Google Scholar] [PubMed]
- Kriegs, M.; Kasten-Pisula, U.; Riepen, B.; Hoffer, K.; Struve, N.; Myllynen, L.; Braig, F.; Binder, M.; Rieckmann, T.; Grénman, R.; et al. Radiosensitization of HNSCC Cells by EGFR Inhibition Depends on the Induction of Cell Cycle Arrests. Oncotarget 2016, 7, 45122–45133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knowles, L.M.; Stabile, L.P.; Egloff, A.M.; Rothstein, M.E.; Thomas, S.M.; Gubish, C.T.; Lerner, E.C.; Seethala, R.R.; Suzuki, S.; Quesnelle, K.M.; et al. HGF and C-Met Participate in Paracrine Tumorigenic Pathways in Head and Neck Squamous Cell Cancer. Clin. Cancer Res. 2009, 15, 3740–3750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.-H.; Lee, J.S.; Kang, S.-O.; Bae, J.-H.; Hong, S.P.; Kahng, H. Serum Hepatocyte Growth Factor as a Marker of Tumor Activity in Head and Neck Squamous Cell Carcinoma. Oral. Oncol. 2007, 43, 1021–1025. [Google Scholar] [CrossRef]
- Kim, I.-G.; Lee, J.-H.; Kim, S.-Y.; Kim, J.-Y.; Cho, E.-W. Fibulin-3 Negatively Regulates ALDH1 via c-MET Suppression and Increases γ-Radiation-Induced Sensitivity in Some Pancreatic Cancer Cell Lines. Biochem. Biophys. Res. Commun. 2014, 454, 369–375. [Google Scholar] [CrossRef] [Green Version]
- Seiwert, T.Y.; Jagadeeswaran, R.; Faoro, L.; Janamanchi, V.; Nallasura, V.; El Dinali, M.; Yala, S.; Kanteti, R.; Cohen, E.E.W.; Lingen, M.W.; et al. The MET Receptor Tyrosine Kinase Is a Potential Novel Therapeutic Target for Head and Neck Squamous Cell Carcinoma. Cancer Res. 2009, 69, 3021–3031. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Li, Q.; Sun, Q.; Zhang, Y.; Yang, H.; Wang, R.; Chen, L.; Wang, W. MET Inhibitor PHA-665752 Suppresses the Hepatocyte Growth Factor-Induced Cell Proliferation and Radioresistance in Nasopharyngeal Carcinoma Cells. Biochem. Biophys. Res. Commun. 2014, 449, 49–54. [Google Scholar] [CrossRef]
- Dy, G.K.; Adjei, A.A. Understanding, Recognizing, and Managing Toxicities of Targeted Anticancer Therapies. CA A Cancer J. Clin. 2013, 63, 249–279. [Google Scholar] [CrossRef]
- Grandis, J.R.; Tweardy, D.J. TGF-Alpha and EGFR in Head and Neck Cancer. J. Cell Biochem. Suppl. 1993, 17F, 188–191. [Google Scholar] [CrossRef]
- Bussink, J.; van der Kogel, A.J.; Kaanders, J.H.A.M. Activation of the PI3-K/AKT Pathway and Implications for Radioresistance Mechanisms in Head and Neck Cancer. Lancet Oncol. 2008, 9, 288–296. [Google Scholar] [CrossRef]
- Medová, M.; Aebersold, D.M.; Zimmer, Y. MET Inhibition in Tumor Cells by PHA665752 Impairs Homologous Recombination Repair of DNA Double Strand Breaks. Int. J. Cancer 2012, 130, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.P.; Franck, D.; Parachoniak, C.A.; Gregg, J.P.; Moore, M.G.; Farwell, D.G.; Rao, S.; Heilmann, A.M.; Erlich, R.L.; Ross, J.S.; et al. MET Genomic Alterations in Head and Neck Squamous Cell Carcinoma (HNSCC): Rapid Response to Crizotinib in a Patient with HNSCC with a Novel MET R1004G Mutation. Oncologist 2019, 24, 1305–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lüttich, L.; Besso, M.J.; Heiden, S.; Koi, L.; Baumann, M.; Krause, M.; Dubrovska, A.; Linge, A.; Kurth, I.; Peitzsch, C. Tyrosine Kinase c-MET as Therapeutic Target for Radiosensitization of Head and Neck Squamous Cell Carcinomas. Cancers 2021, 13, 1865. https://doi.org/10.3390/cancers13081865
Lüttich L, Besso MJ, Heiden S, Koi L, Baumann M, Krause M, Dubrovska A, Linge A, Kurth I, Peitzsch C. Tyrosine Kinase c-MET as Therapeutic Target for Radiosensitization of Head and Neck Squamous Cell Carcinomas. Cancers. 2021; 13(8):1865. https://doi.org/10.3390/cancers13081865
Chicago/Turabian StyleLüttich, Lina, María José Besso, Stephan Heiden, Lydia Koi, Michael Baumann, Mechthild Krause, Anna Dubrovska, Annett Linge, Ina Kurth, and Claudia Peitzsch. 2021. "Tyrosine Kinase c-MET as Therapeutic Target for Radiosensitization of Head and Neck Squamous Cell Carcinomas" Cancers 13, no. 8: 1865. https://doi.org/10.3390/cancers13081865
APA StyleLüttich, L., Besso, M. J., Heiden, S., Koi, L., Baumann, M., Krause, M., Dubrovska, A., Linge, A., Kurth, I., & Peitzsch, C. (2021). Tyrosine Kinase c-MET as Therapeutic Target for Radiosensitization of Head and Neck Squamous Cell Carcinomas. Cancers, 13(8), 1865. https://doi.org/10.3390/cancers13081865