Molecular Assessment of HER2 to Identify Signatures Associated with Therapy Response in HER2-Positive Breast Cancer
Abstract
:Simple Summary
Abstract
1. Background
2. Materials and Methods
2.1. Coverslip Cleaning
2.2. Cell Culture
2.3. Antibodies and Fluorescent Dye Conjugation
2.4. Photophysical Characterization of Fluorescent Probes
2.5. Flow Cytometry in Cultured Cell Lines
2.6. Immunofluorescence Staining of Cultured Cells
2.7. Human Subjects and Core Biopsy Collection
2.8. Tissue Touch Prep and Immunofluorescence Staining
2.9. dSTORM Setup and Image Acquisition
2.10. Image Processing and Analysis
2.11. Statistical Analysis
3. Results
3.1. Molecular Imaging of HER2 in Cultured Breast Cancer Cell Lines
3.2. Quantitative SMLM in luminal B Cultured Breast Cancer Cell Lines
3.3. Quantitative SMLM in HER2-Enriched Breast Cancer Cell Lines
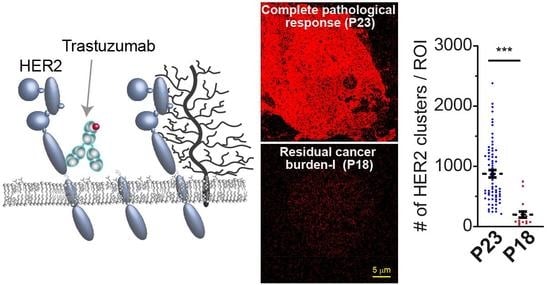
3.4. Detected HER2 Density and Nano-Organization in Patient Tissue Specimens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vogel, C.L.; Cobleigh, M.A.; Tripathy, D.; Gutheil, J.C.; Harris, L.N.; Fehrenbacher, L.; Slamon, D.J.; Murphy, M.; Novotny, W.F.; Burchmore, M.; et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2002, 20, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M.; et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F. HER2 and Breast Cancer—A Phenomenal Success Story. N. Engl. J. Med. 2019, 381, 1284–1286. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 3997–4013. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, T.W.; Gown, A.M.; Yaziji, H.; Barnes, M.J.; Schnitt, S.J. Comparison of fluorescence in situ hybridization and immunohistochemistry for the evaluation of HER-2/neu in breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 1999, 17, 1974–1982. [Google Scholar] [CrossRef] [PubMed]
- Kakar, S.; Puangsuvan, N.; Stevens, J.M.; Serenas, R.; Mangan, G.; Sahai, S.; Mihalov, M.L. HER-2/neu assessment in breast cancer by immunohistochemistry and fluorescence in situ hybridization: Comparison of results and correlation with survival. Mol. Diagn. J. Devoted Underst. Hum. Dis. Clin. Appl. Mol. Biol. 2000, 5, 199–207. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Allison, K.H.; Harvey, B.E.; Mangu, P.B.; Bartlett, J.M.S.; Bilous, M.; Ellis, I.O.; Fitzgibbons, P.; Hanna, W.; et al. Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of american pathologists clinical practice guideline focused update. J. Clin. Oncol. 2018, 36, 2105. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Xu, B. Targeted therapeutic options and future perspectives for HER2-positive breast cancer. Signal. Transduct. Target. Ther. 2019, 4, 34. [Google Scholar] [CrossRef] [Green Version]
- Fehrenbacher, L.; Cecchini, R.S.; Geyer, C.E., Jr.; Rastogi, P.; Costantino, J.P.; Atkins, J.N.; Crown, J.P.; Polikoff, J.; Boileau, J.F.; Provencher, L.; et al. NSABP B-47/NRG oncology phase iii randomized trial comparing adjuvant chemotherapy with or without trastuzumab in high-risk invasive breast cancer negative for HER2 by FISH and with IHC 1+ or 2. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 444–453. [Google Scholar] [CrossRef]
- Piccart-Gebhart, M.J.; Procter, M.; Leyland-Jones, B.; Goldhirsch, A.; Untch, M.; Smith, I.; Gianni, L.; Baselga, J.; Bell, R.; Jackisch, C.; et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N. Engl. J. Med. 2005, 353, 1659–1672. [Google Scholar] [CrossRef] [Green Version]
- Romond, E.H.; Perez, E.A.; Bryant, J.; Suman, V.J.; Geyer, C.E., Jr.; Davidson, N.E.; Tan-Chiu, E.; Martino, S.; Paik, S.; Kaufman, P.A.; et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N. Engl. J. Med. 2005, 353, 1673–1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slamon, D.; Eiermann, W.; Robert, N.; Pienkowski, T.; Martin, M.; Press, M.; Mackey, J.; Glaspy, J.; Chan, A.; Pawlicki, M.; et al. Adjuvant trastuzumab in HER2-positive breast cancer. N. Engl. J. Med. 2011, 365, 1273–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, E.M.; Kostler, W.J.; Horvat, R.; Hudelist, G.; Kubista, E.; Attems, J.; Zielinski, C.C.; Singer, C.F. High-level ERBB2 gene amplification is associated with a particularly short time-to-metastasis, but results in a high rate of complete response once trastuzumab-based therapy is offered in the metastatic setting. Int. J. Cancer J. Int. Du Cancer 2014, 135, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, R.; Durbecq, V.; Di Leo, A.; Paesmans, M.; Larsimont, D.; Leroy, J.Y.; Borms, M.; Vindevoghel, A.; Jerusalem, G.; D'Hondt, V.; et al. Phosphorylated HER-2 tyrosine kinase and Her-2/neu gene amplification as predictive factors of response to trastuzumab in patients with HER-2 overexpressing metastatic breast cancer (MBC). Eur J. Cancer 2007, 43, 725–735. [Google Scholar] [CrossRef]
- Han, H.S.; Kim, J.S.; Park, J.H.; Jeon, Y.K.; Lee, K.W.; Oh, D.Y.; Kim, J.H.; Park, S.Y.; Im, S.A.; Kim, T.Y.; et al. Weekly paclitaxel and trastuzumab as a first-line therapy in patients with HER2-overexpressing metastatic breast cancer: Magnitude of HER2/neu amplification as a predictive factor for efficacy. J. Korean Med. Sci. 2009, 24, 910–917. [Google Scholar] [CrossRef]
- Kim, J.W.; Kim, J.H.; Im, S.A.; Kim, Y.J.; Han, H.S.; Kim, J.S.; Lee, K.H.; Kim, T.Y.; Han, S.W.; Jeon, Y.K.; et al. HER2/CEP17 ratio and HER2 immunohistochemistry predict clinical outcome after first-line trastuzumab plus taxane chemotherapy in patients with HER2 fluorescence in situ hybridization-positive metastatic breast cancer. Cancer Chemother. Pharm. 2013, 72, 109–115. [Google Scholar] [CrossRef]
- Von Minckwitz, G.; Huang, C.S.; Mano, M.S.; Loibl, S.; Mamounas, E.P.; Untch, M.; Wolmark, N.; Rastogi, P.; Schneeweiss, A.; Redondo, A.; et al. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N. Engl. J. Med. 2019, 380, 617–628. [Google Scholar] [CrossRef]
- Jacobs, S.A.; Robidoux, A.; Abraham, J.; Pérez-Garcia, J.M.; La Verde, N.; Orcutt, J.M.; Cazzaniga, M.E.; Piette, F.; Antolín, S.; Aguirre, E.; et al. NSABP FB-7: A phase II randomized neoadjuvant trial with paclitaxel + trastuzumab and/or neratinib followed by chemotherapy and postoperative trastuzumab in HER2+ breast cancer. Breast Cancer Res. 2019, 21, 133. [Google Scholar] [CrossRef]
- Patel, A.; Unni, N.; Peng, Y. The changing paradigm for the treatment of HER2-positive breast cancer. Cancers 2020, 12, 2081. [Google Scholar] [CrossRef]
- FDA/CBER: Clinical Review Briefing Documant sBLA STN:103792Orig1s5311 Trastuzumab. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2014/103792Orig1s5311MedR.pdf (accessed on 18 December 2019).
- Chumsri, S.; Li, Z.; Serie, D.J.; Mashadi-Hossein, A.; Colon-Otero, G.; Song, N.; Pogue-Geile, K.L.; Gavin, P.G.; Paik, S.; Moreno-Aspitia, A.; et al. Incidence of late relapses in patients with HER2-positive breast cancer receiving adjuvant trastuzumab: Combined analysis of NCCTG N9831 (alliance) and NRG Oncology/NSABP B-31. J. Clin. Oncol. 2019, 37, 3425–3435. [Google Scholar] [CrossRef]
- Korde, L.A.; Somerfield, M.R.; Carey, L.A.; Crews, J.R.; Denduluri, N.; Hwang, E.S.; Khan, S.A.; Loibl, S.; Morris, E.A.; Perez, A.; et al. Neoadjuvant chemotherapy, endocrine therapy, and targeted therapy for breast cancer: ASCO guideline. J. Clin. Oncol. 2021, 39, 1485–1505. [Google Scholar] [CrossRef] [PubMed]
- Giordano, S.H.; Temin, S.; Chandarlapaty, S.; Crews, J.R.; Esteva, F.J.; Kirshner, J.J.; Krop, I.E.; Levinson, J.; Lin, N.U.; Modi, S.; et al. Systemic therapy for patients with advanced human epidermal growth factor receptor 2—Positive breast cancer: ASCO clinical practice guideline update. J. Clin. Oncol. 2018, 36, 2736–2740. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, R.; Narasanna, A.; Wang, S.E.; Liu, S.; Chakrabarty, A.; Balko, J.M.; Gonzalez-Angulo, A.M.; Mills, G.B.; Penuel, E.; Winslow, J.; et al. Trastuzumab has preferential activity against breast cancers driven by HER2 homodimers. Cancer Res. 2011, 71, 1871–1882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moasser, M.M. Targeting the function of the HER2 oncogene in human cancer therapeutics. Oncogene 2007, 26, 6577–6592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vu, T.; Claret, F.X. Trastuzumab: Updated mechanisms of action and resistance in breast cancer. Front. Oncol. 2012, 2, 62. [Google Scholar] [CrossRef] [Green Version]
- Molina, M.A.; Codony-Servat, J.; Albanell, J.; Rojo, F.; Arribas, J.; Baselga, J. Trastuzumab (herceptin), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer Res. 2001, 61, 4744–4749. [Google Scholar]
- Spector, N.L.; Blackwell, K.L. Understanding the mechanisms behind trastuzumab therapy for human epidermal growth factor receptor 2-positive breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 5838–5847. [Google Scholar] [CrossRef]
- Dean, L. Trastuzumab (herceptin) therapy and ERBB2 (HER2) genotype. In Medical Genetics Summaries; Pratt, V., McLeod, H., Rubinstein, W., Dean, L., Kattman, B., Malheiro, A., Eds.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2012. [Google Scholar]
- Verma, S.; Miles, D.; Gianni, L.; Krop, I.E.; Welslau, M.; Baselga, J.; Pegram, M.; Oh, D.-Y.; Diéras, V.; Guardino, E.; et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N. Engl. J. Med. 2012, 367, 1783–1791. [Google Scholar] [CrossRef] [Green Version]
- Modi, S.; Saura, C.; Yamashita, T.; Park, Y.H.; Kim, S.-B.; Tamura, K.; Andre, F.; Iwata, H.; Ito, Y.; Tsurutani, J.; et al. Trastuzumab deruxtecan in previously treated HER2-positive breast cancer. N. Engl. J. Med. 2020, 382, 610–621. [Google Scholar] [CrossRef]
- Ogitani, Y.; Aida, T.; Hagihara, K.; Yamaguchi, J.; Ishii, C.; Harada, N.; Soma, M.; Okamoto, H.; Oitate, M.; Arakawa, S.; et al. DS-8201a, a novel HER2-targeting ADC with a novel DNA topoisomerase I inhibitor, demonstrates a promising antitumor efficacy with differentiation from T-DM1. Clin. Cancer Res. 2016, 22, 5097–5108. [Google Scholar] [CrossRef] [Green Version]
- Adams, C.W.; Allison, D.E.; Flagella, K.; Presta, L.; Clarke, J.; Dybdal, N.; McKeever, K.; Sliwkowski, M.X. Humanization of a recombinant monoclonal antibody to produce a therapeutic HER dimerization inhibitor, pertuzumab. Cancer Immunol. Immunother. 2005, 55, 717. [Google Scholar] [CrossRef] [PubMed]
- Tesch, M.E.; Gelmon, K.A. Targeting HER2 in breast cancer: Latest developments on treatment sequencing and the introduction of biosimilars. Drugs 2020, 80, 1811–1830. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Bryant, J.; Tan-Chiu, E.; Yothers, G.; Park, C.; Wickerham, D.L.; Wolmark, N. HER2 and choice of adjuvant chemotherapy for invasive breast cancer: National surgical adjuvant breast and bowel project protocol B-15. JNCI J. Natl. Cancer Inst. 2000, 92, 1991–1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrulis, I.L.; Bull, S.B.; Blackstein, M.E.; Sutherland, D.; Mak, C.; Sidlofsky, S.; Pritzker, K.P.; Hartwick, R.W.; Hanna, W.; Lickley, L.; et al. Neu/erbB-2 amplification identifies a poor-prognosis group of women with node-negative breast cancer. Toronto Breast Cancer Study Group. J. Clin. Oncol. 1998, 16, 1340–1349. [Google Scholar] [CrossRef]
- Seshadri, R.; Firgaira, F.A.; Horsfall, D.J.; McCaul, K.; Setlur, V.; Kitchen, P. Clinical significance of HER-2/neu oncogene amplification in primary breast cancer. The South Australian Breast Cancer Study Group. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 1993, 11, 1936–1942. [Google Scholar] [CrossRef]
- Tandon, A.K.; Clark, G.M.; Chamness, G.C.; Ullrich, A.; McGuire, W.L. HER-2/neu oncogene protein and prognosis in breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 1989, 7, 1120–1128. [Google Scholar] [CrossRef]
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; McGuire, W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Mendes, D.; Alves, C.; Afonso, N.; Cardoso, F.; Passos-Coelho, J.L.; Costa, L.; Andrade, S.; Batel-Marques, F. The benefit of HER2-targeted therapies on overall survival of patients with metastatic HER2-positive breast cancer—A systematic review. Breast Cancer Res. 2015, 17, 140. [Google Scholar] [CrossRef] [Green Version]
- Jackisch, C.; Schoenegg, W.; Reichert, D.; Welslau, M.; Selbach, J.; Harich, H.D.; Tesch, H.; Wohlfarth, T.; Eustermann, H.; Hinke, A. Trastuzumab in advanced breast cancer—A decade of experience in Germany. BMC Cancer 2014, 14, 924. [Google Scholar] [CrossRef] [Green Version]
- de Mattos-Arruda, L.; Cortes, J. Advances in first-line treatment for patients with HER-2+ metastatic breast cancer. Oncologist 2012, 17, 631–644. [Google Scholar] [CrossRef] [Green Version]
- Swain, S.M.; Baselga, J.; Kim, S.B.; Ro, J.; Semiglazov, V.; Campone, M.; Ciruelos, E.; Ferrero, J.M.; Schneeweiss, A.; Heeson, S.; et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N. Engl. J. Med. 2015, 372, 724–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swain, S.M.; Miles, D.; Kim, S.B.; Im, Y.H.; Im, S.A.; Semiglazov, V.; Ciruelos, E.; Schneeweiss, A.; Loi, S.; Monturus, E.; et al. Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA): End-of-study results from a double-blind, randomised, placebo-controlled, phase 3 study. Lancet Oncol. 2020, 21, 519–530. [Google Scholar] [CrossRef]
- Tripathy, D.; Slamon, D.J.; Cobleigh, M.; Arnold, A.; Saleh, M.; Mortimer, J.E.; Murphy, M.; Stewart, S.J. Safety of treatment of metastatic breast cancer with trastuzumab beyond disease progression. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2004, 22, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, R.; Wenzel, C.; Hussian, D.; Pluschnig, U.; Sevelda, U.; Koestler, W.; Altorjai, G.; Locker, G.J.; Mader, R.; Zielinski, C.C.; et al. Analysis of trastuzumab and chemotherapy in advanced breast cancer after the failure of at least one earlier combination: An observational study. BMC Cancer 2006, 6, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sperinde, J.; Jin, X.; Banerjee, J.; Penuel, E.; Saha, A.; Diedrich, G.; Huang, W.; Leitzel, K.; Weidler, J.; Ali, S.M.; et al. Quantitation of p95HER2 in paraffin sections by using a p95-specific antibody and correlation with outcome in a cohort of trastuzumab-treated breast cancer patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 4226–4235. [Google Scholar] [CrossRef] [Green Version]
- Scaltriti, M.; Rojo, F.; Ocana, A.; Anido, J.; Guzman, M.; Cortes, J.; Di Cosimo, S.; Matias-Guiu, X.; Ramon y Cajal, S.; Arribas, J.; et al. Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J. Natl. Cancer Inst. 2007, 99, 628–638. [Google Scholar] [CrossRef] [Green Version]
- Pedersen, K.; Angelini, P.D.; Laos, S.; Bach-Faig, A.; Cunningham, M.P.; Ferrer-Ramon, C.; Luque-Garcia, A.; Garcia-Castillo, J.; Parra-Palau, J.L.; Scaltriti, M.; et al. A naturally occurring HER2 carboxy-terminal fragment promotes mammary tumor growth and metastasis. Mol. Cell. Biol. 2009, 29, 3319–3331. [Google Scholar] [CrossRef] [Green Version]
- Saez, R.; Molina, M.A.; Ramsey, E.E.; Rojo, F.; Keenan, E.J.; Albanell, J.; Lluch, A.; Garcia-Conde, J.; Baselga, J.; Clinton, G.M. P95HER-2 predicts worse outcome in patients with HER-2-positive breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 424–431. [Google Scholar] [CrossRef] [Green Version]
- Nagy, P.; Friedlander, E.; Tanner, M.; Kapanen, A.I.; Carraway, K.L.; Isola, J.; Jovin, T.M. Decreased accessibility and lack of activation of ErbB2 in JIMT-1, a herceptin-resistant, MUC4-expressing breast cancer cell line. Cancer Res. 2005, 65, 473–482. [Google Scholar]
- Martin-Castillo, B.; Oliveras-Ferraros, C.; Vazquez-Martin, A.; Cufí, S.; Moreno, J.M.; Corominas-Faja, B.; Urruticoechea, A.; Martín, Á.G.; López-Bonet, E.; Menendez, J.A. Basal/HER2 breast carcinomas: Integrating molecular taxonomy with cancer stem cell dynamics to predict primary resistance to trastuzumab (Herceptin). Cell Cycle 2013, 12, 225–245. [Google Scholar] [CrossRef] [Green Version]
- Ng, C.K.Y.; Bidard, F.C.; Piscuoglio, S.; Geyer, F.C.; Lim, R.S.; de Bruijn, I.; Shen, R.; Pareja, F.; Berman, S.H.; Wang, L.; et al. Genetic heterogeneity in therapy-naïve synchronous primary breast cancers and their metastases. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 4402–4415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yates, L.R.; Gerstung, M.; Knappskog, S.; Desmedt, C.; Gundem, G.; Van Loo, P.; Aas, T.; Alexandrov, L.B.; Larsimont, D.; Davies, H.; et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat. Med. 2015, 21, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Zardavas, D.; Irrthum, A.; Swanton, C.; Piccart, M. Clinical management of breast cancer heterogeneity. Nat. Rev. Clin. Oncol. 2015, 12, 381–394. [Google Scholar] [CrossRef] [PubMed]
- Balko, J.M.; Giltnane, J.M.; Wang, K.; Schwarz, L.J.; Young, C.D.; Cook, R.S.; Owens, P.; Sanders, M.E.; Kuba, M.G.; Sánchez, V.; et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014, 4, 232–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branco, F.P.; Machado, D.; Silva, F.F.; André, S.; Catarino, A.; Madureira, R.; Pinto, J.M.; Godinho, J.P.; Simões, P.D.; Brito, M.; et al. Loss of HER2 and disease prognosis after neoadjuvant treatment of HER2+ breast cancer. Am. J. Transl. Res. 2019, 11, 6110–6116. [Google Scholar]
- Ritter, C.A.; Perez-Torres, M.; Rinehart, C.; Guix, M.; Dugger, T.; Engelman, J.A.; Arteaga, C.L. Human breast cancer cells selected for resistance to trastuzumab in vivo overexpress epidermal growth factor receptor and ErbB ligands and remain dependent on the ErbB receptor network. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 4909–4919. [Google Scholar] [CrossRef] [Green Version]
- Harris, L.N.; You, F.; Schnitt, S.J.; Witkiewicz, A.; Lu, X.; Sgroi, D.; Ryan, P.D.; Come, S.E.; Burstein, H.J.; Lesnikoski, B.A.; et al. Predictors of resistance to preoperative trastuzumab and vinorelbine for HER2-positive early breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2007, 13, 1198–1207. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J.; Seo, A.N.; Kim, E.J.; Jang, M.H.; Kim, Y.J.; Kim, J.H.; Kim, S.W.; Ryu, H.S.; Park, I.A.; Im, S.A.; et al. Prognostic and predictive values of EGFR overexpression and EGFR copy number alteration in HER2-positive breast cancer. Br. J. Cancer 2015, 112, 103–111. [Google Scholar] [CrossRef] [Green Version]
- Gala, K.; Chandarlapaty, S. Molecular pathways: HER3 targeted therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 1410–1416. [Google Scholar] [CrossRef] [Green Version]
- Cheng, H.; Ballman, K.; Vassilakopoulou, M.; Dueck, A.C.; Reinholz, M.M.; Tenner, K.; Gralow, J.; Hudis, C.; Davidson, N.E.; Fountzilas, G.; et al. EGFR expression is associated with decreased benefit from trastuzumab in the NCCTG N9831 (Alliance) trial. Br. J. Cancer 2014, 111, 1065–1071. [Google Scholar] [CrossRef] [Green Version]
- Gallardo, A.; Lerma, E.; Escuin, D.; Tibau, A.; Munoz, J.; Ojeda, B.; Barnadas, A.; Adrover, E.; Sanchez-Tejada, L.; Giner, D.; et al. Increased signalling of EGFR and IGF1R, and deregulation of PTEN/PI3K/Akt pathway are related with trastuzumab resistance in HER2 breast carcinomas. Br. J. Cancer 2012, 106, 1367–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Zi, X.; Zhao, Y.; Mascarenhas, D.; Pollak, M. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin). J. Natl. Cancer Inst. 2001, 93, 1852–1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nahta, R.; Yuan, L.X.; Zhang, B.; Kobayashi, R.; Esteva, F.J. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005, 65, 11118–11128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luque-Cabal, M.; Garcia-Teijido, P.; Fernandez-Perez, Y.; Sanchez-Lorenzo, L.; Palacio-Vazquez, I. Mechanisms behind the resistance to trastuzumab in HER2-amplified breast cancer and strategies to overcome it. Clin. Med. Insights Oncol. 2016, 10, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Jerome, L.; Alami, N.; Belanger, S.; Page, V.; Yu, Q.; Paterson, J.; Shiry, L.; Pegram, M.; Leyland-Jones, B. Recombinant human insulin-like growth factor binding protein 3 inhibits growth of human epidermal growth factor receptor-2-overexpressing breast tumors and potentiates herceptin activity in vivo. Cancer Res. 2006, 66, 7245–7252. [Google Scholar] [CrossRef] [Green Version]
- Oliveras-Ferraros, C.; Vazquez-Martin, A.; Martin-Castillo, B.; Perez-Martinez, M.C.; Cufi, S.; Del Barco, S.; Bernado, L.; Brunet, J.; Lopez-Bonet, E.; Menendez, J.A. Pathway-focused proteomic signatures in HER2-overexpressing breast cancer with a basal-like phenotype: New insights into de novo resistance to trastuzumab (Herceptin). Int. J. Oncol. 2010, 37, 669–678. [Google Scholar] [PubMed]
- Nahta, R. Deciphering the role of insulin-like growth factor-I receptor in trastuzumab resistance. Chemother. Res. Pract. 2012, 2012, 648965. [Google Scholar] [CrossRef] [Green Version]
- Corcoran, C.; Rani, S.; Breslin, S.; Gogarty, M.; Ghobrial, I.M.; Crown, J.; O'Driscoll, L. MiR-630 targets IGF1R to regulate response to HER-targeting drugs and overall cancer cell progression in HER2 over-expressing breast cancer. Mol. Cancer 2014, 13, 71. [Google Scholar] [CrossRef] [Green Version]
- Alexander, P.B.; Chen, R.; Gong, C.; Yuan, L.; Jasper, J.S.; Ding, Y.; Markowitz, G.J.; Yang, P.; Xu, X.; McDonnell, D.P.; et al. Distinct receptor tyrosine kinase subsets mediate anti-HER2 drug resistance in breast cancer. J. Biol Chem. 2017, 292, 748–759. [Google Scholar] [CrossRef] [Green Version]
- Weitsman, G.; Barber, P.R.; Nguyen, L.K.; Lawler, K.; Patel, G.; Woodman, N.; Kelleher, M.T.; Pinder, S.E.; Rowley, M.; Ellis, P.A.; et al. HER2-HER3 dimer quantification by FLIM-FRET predicts breast cancer metastatic relapse independently of HER2 IHC status. Oncotarget 2016, 7, 51012–51026. [Google Scholar] [CrossRef] [Green Version]
- Amin, D.N.; Campbell, M.R.; Moasser, M.M. The role of HER3, the unpretentious member of the HER family, in cancer biology and cancer therapeutics. Semin. Cell Dev. Biol. 2010, 21, 944–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sergina, N.V.; Moasser, M.M. The HER family and cancer: Emerging molecular mechanisms and therapeutic targets. Trends Mol. Med. 2007, 13, 527–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berns, K.; Horlings, H.M.; Hennessy, B.T.; Madiredjo, M.; Hijmans, E.M.; Beelen, K.; Linn, S.C.; Gonzalez-Angulo, A.M.; Stemke-Hale, K.; Hauptmann, M.; et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 2007, 12, 395–402. [Google Scholar] [CrossRef] [Green Version]
- Nagata, Y.; Lan, K.H.; Zhou, X.; Tan, M.; Esteva, F.J.; Sahin, A.A.; Klos, K.S.; Li, P.; Monia, B.P.; Nguyen, N.T.; et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 2004, 6, 117–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kataoka, Y.; Mukohara, T.; Shimada, H.; Saijo, N.; Hirai, M.; Minami, H. Association between gain-of-function mutations in PIK3CA and resistance to HER2-targeted agents in HER2-amplified breast cancer cell lines. Ann. Oncol. 2010, 21, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.E.; Ferraro, E.; Safonov, A.; Morales, C.B.; Lahuerta, E.J.A.; Li, Q.; Kulick, A.; Ross, D.; Solit, D.B.; de Stanchina, E.; et al. HER2 + breast cancers evade anti-HER2 therapy via a switch in driver pathway. Nat. Commun. 2021, 12, 6667. [Google Scholar] [CrossRef] [PubMed]
- Nahta, R.; Takahashi, T.; Ueno, N.T.; Hung, M.C.; Esteva, F.J. P27(kip1) down-regulation is associated with trastuzumab resistance in breast cancer cells. Cancer Res. 2004, 64, 3981–3986. [Google Scholar] [CrossRef] [Green Version]
- Obr, A.E.; Kumar, S.; Chang, Y.-J.; Bulatowicz, J.J.; Barnes, B.J.; Birge, R.B.; Lazzarino, D.A.; Gallagher, E.; LeRoith, D.; Wood, T.L. Insulin-like growth factor receptor signaling in breast tumor epithelium protects cells from endoplasmic reticulum stress and regulates the tumor microenvironment. Breast Cancer Res. BCR 2018, 20, 138. [Google Scholar] [CrossRef]
- McDermott, M.S.J.; Canonici, A.; Ivers, L.; Browne, B.C.; Madden, S.F.; O'Brien, N.A.; Crown, J.; O’Donovan, N. Dual inhibition of IGF1R and ER enhances response to trastuzumab in HER2 positive breast cancer cells. Int. J. Oncol. 2017, 50, 2221–2228. [Google Scholar] [CrossRef] [Green Version]
- Ekyalongo, R.C.; Yee, D. Revisiting the IGF-1R as a breast cancer target. NPJ Precis. Oncol. 2017, 1, 14. [Google Scholar] [CrossRef] [Green Version]
- Sanabria-Figueroa, E.; Donnelly, S.M.; Foy, K.C.; Buss, M.C.; Castellino, R.C.; Paplomata, E.; Taliaferro-Smith, L.; Kaumaya, P.T.; Nahta, R. Insulin-like growth factor-1 receptor signaling increases the invasive potential of human epidermal growth factor receptor 2-overexpressing breast cancer cells via Src-focal adhesion kinase and forkhead box protein M1. Mol. Pharm. 2015, 87, 150–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhargava, R.; Beriwal, S.; McManus, K.; Dabbs, D.J. Insulin-like growth factor receptor-1 (IGF-1R) expression in normal breast, proliferative breast lesions, and breast carcinoma. Appl. Immunohistochem. Mol. Morphol. AIMM 2011, 19, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Browne, B.C.; Crown, J.; Venkatesan, N.; Duffy, M.J.; Clynes, M.; Slamon, D.; O'Donovan, N. Inhibition of IGF1R activity enhances response to trastuzumab in HER-2-positive breast cancer cells. Ann. Oncol. 2010, 22, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Lee-Hoeflich, S.T.; Crocker, L.; Yao, E.; Pham, T.; Munroe, X.; Hoeflich, K.P.; Sliwkowski, M.X.; Stern, H.M. A central role for HER3 in HER2-amplified breast cancer: Implications for targeted therapy. Cancer Res. 2008, 68, 5878–5887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fehling-Kaschek, M.; Peckys, D.B.; Kaschek, D.; Timmer, J.; Jonge, N.d. Mathematical modeling of drug-induced receptor internalization in the HER2-positive SKBR3 breast cancer cell-line. Sci. Rep. 2019, 9, 12709. [Google Scholar] [CrossRef] [Green Version]
- Rappoport, J.Z.; Simon, S.M. Endocytic trafficking of activated EGFR is AP-2 dependent and occurs through preformed clathrin spots. J. Cell Sci. 2009, 122, 1301–1305. [Google Scholar] [CrossRef] [Green Version]
- Kast, F.; Schwill, M.; Stüber, J.C.; Pfundstein, S.; Nagy-Davidescu, G.; Rodríguez, J.M.M.; Seehusen, F.; Richter, C.P.; Honegger, A.; Hartmann, K.P.; et al. Engineering an anti-HER2 biparatopic antibody with a multimodal mechanism of action. Nat. Commun. 2021, 12, 3790. [Google Scholar] [CrossRef]
- Stüber, J.C.; Richter, C.P.; Bellón, J.S.; Schwill, M.; König, I.; Schuler, B.; Piehler, J.; Plückthun, A. Apoptosis-inducing anti-HER2 agents operate through oligomerization-induced receptor immobilization. Commun. Biol. 2021, 4, 762. [Google Scholar] [CrossRef]
- Wymant, J.M.; Sayers, E.J.; Muir, D.; Jones, A.T. Strategic Trastuzumab Mediated Crosslinking Driving Concomitant HER2 and HER3 Endocytosis and Degradation in Breast Cancer. J. Cancer 2020, 11, 3288–3302. [Google Scholar] [CrossRef]
- Dahmke, I.N.; Trampert, P.; Weinberg, F.; Mostajeran, Z.; Lautenschläger, F.; de Jonge, N. Correlative Fluorescence- and Electron Microscopy of Whole Breast Cancer Cells Reveals Different Distribution of ErbB2 Dependent on Underlying Actin. Front. Cell Dev. Biol. 2020, 8, 521. [Google Scholar] [CrossRef]
- Ilgen, P.; Stoldt, S.; Conradi, L.C.; Wurm, C.A.; Ruschoff, J.; Ghadimi, B.M.; Liersch, T.; Jakobs, S. STED super-resolution microscopy of clinical paraffin-embedded human rectal cancer tissue. PLoS ONE 2014, 9, e101563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Creech, M.K.; Wang, J.; Nan, X.; Gibbs, S.L. Superresolution imaging of clinical formalin fixed paraffin embedded breast cancer with single molecule localization microscopy. Sci. Rep. 2017, 7, 40766. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yao, L.; Jing, Y.; Fei, Y.; Bai, Q.; Mi, L.; Ma, J. Multicomposite super-resolution microscopy: Enhanced airyscan resolution with radial fluctuation and sample expansions. J. Biophotonics 2020, 13, e2419. [Google Scholar] [CrossRef] [PubMed]
- Golfetto, O.; Wakefield, D.L.; Cacao, E.E.; Avery, K.N.; Kenyon, V.; Jorand, R.; Tobin, S.J.; Biswas, S.; Gutierrez, J.; Clinton, R.; et al. A platform to enhance quantitative single molecule localization microscopy. J. Am. Chem. Soc. 2018, 140, 12785–12797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tobin, S.J.; Wakefield, D.L.; Liu, X.; Jones, V.; Schmolze, D.; Jovanovic-Talisman, T. Single molecule localization microscopy coupled with touch preparation for the quantification of trastuzumab-bound HER2. Sci. Rep. 2018, 11, 15154. [Google Scholar] [CrossRef] [Green Version]
- Jorand, R.; Biswas, S.; Wakefield, D.L.; Tobin, S.J.; Golfetto, O.; Hilton, K.; Ko, M.; Ramos, J.W.; Small, A.R.; Chu, P.; et al. Molecular signatures of mu opioid receptor and somatostatin receptor 2 in pancreatic cancer. Mol. Biol. Cell 2016, 27, 3659–3672. [Google Scholar] [CrossRef] [Green Version]
- Kute, T.; Lack, C.M.; Willingham, M.; Bishwokama, B.; Williams, H.; Barrett, K.; Mitchell, T.; Vaughn, J.P. Development of herceptin resistance in breast cancer cells. Cytom. Part A 2004, 57A, 86–93. [Google Scholar] [CrossRef]
- Dempsey, G.T.; Vaughan, J.C.; Chen, K.H.; Bates, M.; Zhuang, X.W. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 2011, 8, 1027–1036. [Google Scholar] [CrossRef]
- Annibale, P.; Vanni, S.; Scarselli, M.; Rothlisberger, U.; Radenovic, A. Identification of clustering artifacts in photoactivated localization microscopy. Nat. Methods 2011, 8, 527–528. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, P.; Jovanovic-Talisman, T.; Skoko, D.; Renz, M.; Veatch, S.L.; Lippincott-Schwartz, J. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat. Methods 2011, 8, 969–975. [Google Scholar] [CrossRef]
- Sengupta, P.; Jovanovic-Talisman, T.; Lippincott-Schwartz, J. Quantifying spatial organization in point-localization superresolution images using pair correlation analysis. Nat. Protoc. 2013, 8, 345–354. [Google Scholar] [CrossRef] [Green Version]
- Subik, K.; Lee, J.-F.; Baxter, L.; Strzepek, T.; Costello, D.; Crowley, P.; Xing, L.; Hung, M.-C.; Bonfiglio, T.; Hicks, D.G.; et al. The expression patterns of eR, pR, HeR2, cK5/6, eGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer Basic Clin. Res. 2010, 4, 35–41. [Google Scholar] [CrossRef]
- Palyi-Krekk, Z.; Barok, M.; Isola, J.; Tammi, M.; Szollosi, J.; Nagy, P. Hyaluronan-induced masking of ErbB2 and CD44-enhanced trastuzumab internalisation in trastuzumab resistant breast cancer. Eur. J. Cancer 2007, 43, 2423–2433. [Google Scholar] [CrossRef] [Green Version]
- Capelan, M.; Pugliano, L.; De Azambuja, E.; Bozovic, I.; Saini, K.S.; Sotiriou, C.; Loi, S.; Piccart-Gebhart, M.J. Pertuzumab: New hope for patients with HER2-positive breast cancer. Ann. Oncol. 2013, 24, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Metzger-Filho, O.; Winer, E.P.; Krop, I. Pertuzumab: Optimizing HER2 blockade. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 5552–5556. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.A.K.; Suzuki, K.G.N.; Shirai, Y.M.; Shibutani, S.T.; Miyahara, M.S.H.; Tsuboi, H.; Yahara, M.; Yoshimura, A.; Mayor, S.; Fujiwara, T.K.; et al. Membrane molecules mobile even after chemical fixation. Nat. Methods 2010, 7, 865–866. [Google Scholar] [CrossRef]
- Wakefield, D.L.; Tobin, S.J.; Schmolze, D.; Jovanovic-Talisman, T. Molecular Imaging of HER2 in Patient Tissues with Touch Prep-Quantitative Single Molecule Localization Microscopy; Springer: New York, NY, USA, 2021; Volume 15. [Google Scholar]
- Rubio, I.T.; Korourian, S.; Cowan, C.; Krag, D.N.; Colvert, M.; Klimberg, V.S. Use of touch preps for intraoperative diagnosis of sentinel lymph node metastases in breast cancer. Ann. Surg. Oncol. 1998, 5, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, B.S.; Werb, Z. Stromal effects on mammary gland development and breast cancer. Science 2002, 296, 1046–1049. [Google Scholar] [CrossRef] [Green Version]
- Hamy, A.-S.; Darrigues, L.; Laas, E.; de Croze, D.; Topciu, L.; Lam, G.-T.; Evrevin, C.; Rozette, S.; Laot, L.; Lerebours, F.; et al. Prognostic value of the residual cancer burden index according to breast cancer subtype: Validation on a cohort of BC patients treated by neoadjuvant chemotherapy. PLoS ONE 2020, 15, e0234191. [Google Scholar] [CrossRef]
- Yau, C.; Osdoit, M.; van der Noordaa, M.; Shad, S.; Wei, J.; de Croze, D.; Hamy, A.S.; Lae, M.; Reyal, F.; Sonke, G.S.; et al. Residual cancer burden after neoadjuvant chemotherapy and long-term survival outcomes in breast cancer: A multi-center pooled analysis of 5161 patients. Lancet Oncol. 2022, 23, 23,149–160. [Google Scholar] [CrossRef]
- Hommelgaard, A.M.; Lerdrup, M.; van Deurs, B. Association with membrane protrusions makes ErbB2 an internalization-resistant receptor. Mol. Biol. Cell 2004, 15, 1557–1567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanley, A.; Ashrafi, G.H.; Seddon, A.M.; Modjtahedi, H. Synergistic effects of various Her inhibitors in combination with IGF-1R, C-MET and Src targeting agents in breast cancer cell lines. Sci. Rep. 2017, 7, 3964. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Mohan, N.; Nussinov, R.; Ma, B.; Wu, W.J. Trastuzumab Blocks the receiver function of HER2 leading to the population shifts of HER2-containing homodimers and heterodimers. Antibodies 2021, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Cancer Cell Line Encyclopedia. Available online: https://data.broadinstitute.org/ccle_legacy_data/dna_copy_number/CCLE_copynumber_byGene_2013-12-03.txt (accessed on 28 April 2022).
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Mercogliano, M.F.; De Martino, M.; Venturutti, L.; Rivas, M.A.; Proietti, C.J.; Inurrigarro, G.; Frahm, I.; Allemand, D.H.; Deza, E.G.; Ares, S.; et al. TNFalpha-Induced mucin 4 expression elicits trastuzumab resistance in HER2-positive breast cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 636–648. [Google Scholar] [CrossRef] [Green Version]
- Bear, H.D.; Anderson, S.; Smith, R.E.; Geyer, C.E., Jr.; Mamounas, E.P.; Fisher, B.; Brown, A.M.; Robidoux, A.; Margolese, R.; Kahlenberg, M.S.; et al. Sequential preoperative or postoperative docetaxel added to preoperative doxorubicin plus cyclophosphamide for operable breast cancer: National surgical adjuvant breast and bowel project protocol B-27. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2006, 24, 2019–2027. [Google Scholar] [CrossRef]
- Bear, H.D.; Tang, G.; Rastogi, P.; Geyer, C.E., Jr.; Robidoux, A.; Atkins, J.N.; Baez-Diaz, L.; Brufsky, A.M.; Mehta, R.S.; Fehrenbacher, L.; et al. Bevacizumab added to neoadjuvant chemotherapy for breast cancer. N. Engl. J. Med. 2012, 366, 310–320. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maddox, A.L.; Brehove, M.S.; Eliato, K.R.; Saftics, A.; Romano, E.; Press, M.F.; Mortimer, J.; Jones, V.; Schmolze, D.; Seewaldt, V.L.; et al. Molecular Assessment of HER2 to Identify Signatures Associated with Therapy Response in HER2-Positive Breast Cancer. Cancers 2022, 14, 2795. https://doi.org/10.3390/cancers14112795
Maddox AL, Brehove MS, Eliato KR, Saftics A, Romano E, Press MF, Mortimer J, Jones V, Schmolze D, Seewaldt VL, et al. Molecular Assessment of HER2 to Identify Signatures Associated with Therapy Response in HER2-Positive Breast Cancer. Cancers. 2022; 14(11):2795. https://doi.org/10.3390/cancers14112795
Chicago/Turabian StyleMaddox, Adam L., Matthew S. Brehove, Kiarash R. Eliato, Andras Saftics, Eugenia Romano, Michael F. Press, Joanne Mortimer, Veronica Jones, Daniel Schmolze, Victoria L. Seewaldt, and et al. 2022. "Molecular Assessment of HER2 to Identify Signatures Associated with Therapy Response in HER2-Positive Breast Cancer" Cancers 14, no. 11: 2795. https://doi.org/10.3390/cancers14112795
APA StyleMaddox, A. L., Brehove, M. S., Eliato, K. R., Saftics, A., Romano, E., Press, M. F., Mortimer, J., Jones, V., Schmolze, D., Seewaldt, V. L., & Jovanovic-Talisman, T. (2022). Molecular Assessment of HER2 to Identify Signatures Associated with Therapy Response in HER2-Positive Breast Cancer. Cancers, 14(11), 2795. https://doi.org/10.3390/cancers14112795