Circular RNA hsa_circ_0062682 Binds to YBX1 and Promotes Oncogenesis in Hepatocellular Carcinoma
Abstract
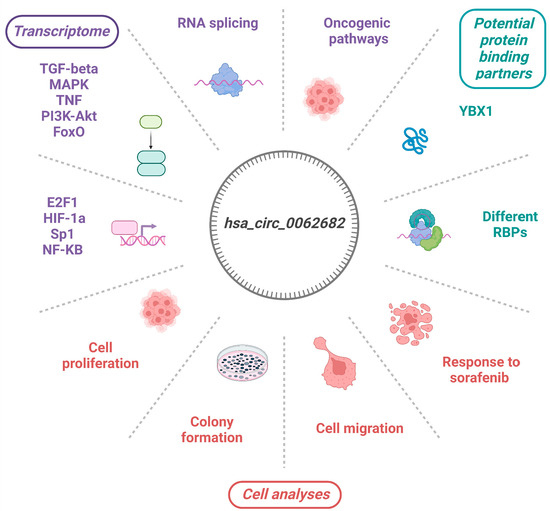
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Differentially Expressed circRNAs in HCC Tumours
2.2. Cell Lines
2.3. Construction of a Modified Mini-Gene System for CircRNA Overexpression
2.4. Transient Transfection
2.5. Generation of Stable Cell Lines
2.6. RNase R Treatment
2.7. Cytoplasmic and Nuclear Localization of RNA and Proteins
2.8. Actinomycin D Treatment
2.9. Proliferation Assay
2.10. Wound Healing Assay
2.11. Invasion Assay
2.12. Colony Forming Assay
2.13. EdU Assay
2.14. Immunostaining
2.15. Sorafenib Treatment
2.16. Biotinylated Oligonucleotide Pulldown
2.17. RNA Immunoprecipitation (RIP)
2.18. RNA Isolation, Reverse Transcription and RT-qPCR
2.19. Western Blotting
2.20. Silver Staining
2.21. Mass Spectrometry
2.22. Functional Protein Network Analysis
2.23. Identification of Differentially Expressed Genes in HCC
2.24. Survival Analysis
2.25. Gene Expression Profiling, Gene Set Enrichment Analysis and Pathway Enrichment Analysis
2.26. Statistical Analysis
3. Results
3.1. CircRNA hsa_circ_0062682 Is Upregulated in Hepatocellular Carcinoma
3.2. Perturbation of hsa_circ_0062682 Affected the Oncogenic Potential of HCC Cell Line Models
3.3. Modulation of the hsa_circ_0062682 Expression Induced Changes in the Transcriptome of HCC Model Cell Lines
3.4. Hsa_circ_0062682 Binds to YBX1 and Its Effect Is Cell-Type Specific
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, A.; Lee, F.-C. Medical Oncology Management of Advanced Hepatocellular Carcinoma 2019: A Reality Check. Front. Med. 2020, 14, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Fitzmaurice, C.; Abate, D.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdel-Rahman, O.; Abdelalim, A.; Abdoli, A.; Abdollahpour, I.; Abdulle, A.S.M.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular Carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-S.; Heo, J.; Libbrecht, L.; Chu, I.-S.; Kaposi-Novak, P.; Calvisi, D.F.; Mikaelyan, A.; Roberts, L.R.; Demetris, A.J.; Sun, Z.; et al. A Novel Prognostic Subtype of Human Hepatocellular Carcinoma Derived from Hepatic Progenitor Cells. Nat. Med. 2006, 12, 410–416. [Google Scholar] [CrossRef]
- Boyault, S.; Rickman, D.S.; de Reyniès, A.; Balabaud, C.; Rebouissou, S.; Jeannot, E.; Hérault, A.; Saric, J.; Belghiti, J.; Franco, D.; et al. Transcriptome Classification of HCC Is Related to Gene Alterations and to New Therapeutic Targets. Hepatology 2007, 45, 42–52. [Google Scholar] [CrossRef]
- Chiang, D.Y.; Villanueva, A.; Hoshida, Y.; Peix, J.; Newell, P.; Minguez, B.; LeBlanc, A.C.; Donovan, D.J.; Thung, S.N.; Solé, M.; et al. Focal Gains of VEGFA and Molecular Classification of Hepatocellular Carcinoma. Cancer Res. 2008, 68, 6779–6788. [Google Scholar] [CrossRef]
- Hoshida, Y.; Nijman, S.M.B.; Kobayashi, M.; Chan, J.A.; Brunet, J.-P.; Chiang, D.Y.; Villanueva, A.; Newell, P.; Ikeda, K.; Hashimoto, M.; et al. Integrative Transcriptome Analysis Reveals Common Molecular Subclasses of Human Hepatocellular Carcinoma. Cancer Res. 2009, 69, 7385–7392. [Google Scholar] [CrossRef]
- Ally, A.; Balasundaram, M.; Carlsen, R.; Chuah, E.; Clarke, A.; Dhalla, N.; Holt, R.A.; Jones, S.J.M.; Lee, D.; Ma, Y.; et al. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017, 169, 1327–1341. [Google Scholar] [CrossRef]
- Caruso, S.; Calatayud, A.L.; Pilet, J.; La Bella, T.; Rekik, S.; Imbeaud, S.; Letouzé, E.; Meunier, L.; Bayard, Q.; Rohr-Udilova, N.; et al. Analysis of Liver Cancer Cell Lines Identifies Agents With Likely Efficacy Against Hepatocellular Carcinoma and Markers of Response. Gastroenterology 2019, 157, 760–776. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The Biogenesis, Biology and Characterization of Circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y. Circular RNAs in Hepatocellular Carcinoma: Emerging Functions to Clinical Significances. Front. Oncol. 2021, 11, 1–17. [Google Scholar] [CrossRef]
- Stagsted, L.V.W.; O’leary, E.T.; Ebbesen, K.K.; Hansen, T.B. The Rna-Binding Protein Sfpq Preserves Long-Intron Splicing and Regulates Circrna Biogenesis in Mammals. Elife 2021, 10, e63088. [Google Scholar] [CrossRef]
- Shen, H.; An, O.; Ren, X.; Song, Y.; Tang, S.J.; Ke, X.-Y.; Han, J.; Tay, D.J.T.; Ng, V.H.E.; Molias, F.B.; et al. ADARs Act as Potent Regulators of Circular Transcriptome in Cancer. Nat. Commun. 2022, 13, 1508. [Google Scholar] [CrossRef]
- Kokot, K.E.; Kneuer, J.M.; John, D.; Rebs, S.; Möbius-Winkler, M.N.; Erbe, S.; Müller, M.; Andritschke, M.; Gaul, S.; Sheikh, B.N.; et al. Reduction of A-to-I RNA Editing in the Failing Human Heart Regulates Formation of Circular RNAs. Basic Res. Cardiol. 2022, 117, 32. [Google Scholar] [CrossRef]
- Razpotnik, R.; Nassib, P.; Kunej, T.; Rozman, D.; Režen, T. Identification of Novel RNA Binding Proteins Influencing Circular RNA Expression in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 7477. [Google Scholar] [CrossRef]
- Louis, C.; Leclerc, D.; Coulouarn, C. Emerging Roles of Circular RNAs in Liver Cancer. JHEP Rep. 2022, 4, 100413. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets-Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Fu, L.; Yao, T.; Chen, Q.; Mo, X.; Hu, Y.; Guo, J. Screening Differential Circular RNA Expression Profiles Reveals Hsa_circ_0004018 Is Associated with Hepatocellular Carcinoma. Oncotarget 2017, 8, 58405–58416. [Google Scholar] [CrossRef]
- Han, D.; Li, J.; Wang, H.; Su, X.; Hou, J.; Gu, Y.; Qian, C.; Lin, Y.; Liu, X.; Huang, M.; et al. Circular RNA CircMTO1 Acts as the Sponge of MicroRNA-9 to Suppress Hepatocellular Carcinoma Progression. Hepatology 2017, 66, 1151–1164. [Google Scholar] [CrossRef]
- Kramer, M.C.; Liang, D.; Tatomer, D.C.; Gold, B.; March, Z.M.; Cherry, S.; Wilusz, J.E. Combinatorial Control of Drosophila Circular RNA Expression by Intronic Repeats, HnRNPs, and SR Proteins. Genes Dev. 2015, 29, 2168–2182. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, E.; Harshman, K.; Kemler, I.; Malipiero, U.; Schaffner, W.; Fontana, A. Astrocytes and Glioblastoma Cells Express Novel Octamer-DNA Binding Proteins Distinct from the Ubiquitous Oct-1 and B Cell Type Oct-2 Proteins. Nucleic Acids Res. 1990, 18, 5495–5503. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, J.; Deng, F.; Yan, Z.; Xia, Y.; Wang, Z.; Ye, J.; Deng, Y.; Zhang, Z.; Qiao, M.; et al. TqPCR: A Touchdown QPCR Assay with Significantly Improved Detection Sensitivity and Amplification Efficiency of SYBR Green QPCR. PLoS ONE 2015, 10, e0132666. [Google Scholar] [CrossRef]
- Gharahdaghi, F.; Weinberg, C.R.; Meagher, D.A.; Imai, B.S.; Mische, S.M. Mass Spectrometric Identification of Proteins from Silver-Stained Polyacrylamide Gel: A Method for the Removal of Silver Ions to Enhance Sensitivity. Electrophoresis 1999, 20, 601–605. [Google Scholar] [CrossRef]
- Keller, A.; Nesvizhskii, A.I.; Kolker, E.; Aebersold, R. Empirical Statistical Model to Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal. Chem. 2002, 74, 5383–5392. [Google Scholar] [CrossRef]
- Nesvizhskii, A.I.; Keller, A.; Kolker, E.; Aebersold, R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003, 75, 4646–4658. [Google Scholar] [CrossRef]
- Mellacheruvu, D.; Wright, Z.; Couzens, A.L.; Lambert, J.-P.; St-Denis, N.A.; Li, T.; Miteva, Y.V.; Hauri, S.; Sardiu, M.E.; Low, T.Y.; et al. The CRAPome: A Contaminant Repository for Affinity Purification-Mass Spectrometry Data. Nat. Methods 2013, 10, 730–736. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor Package for Integrative Analysis of TCGA Data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Menyhárt, O.; Nagy, Á.; Gyórffy, B. Determining Consistent Prognostic Biomarkers of Overall Survival and Vascular Invasion in Hepatocellular Carcinoma. R. Soc. Open Sci. 2018, 5, 181006. [Google Scholar] [CrossRef]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway Enrichment Analysis and Visualization of Omics Data Using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-Type Specific Features of Circular RNA Expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Giannelli, G.; Villa, E.; Lahn, M. Transforming Growth Factor-β as a Therapeutic Target in Hepatocellular Carcinoma. Cancer Res. 2014, 74, 1890–1894. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gingold, J.A.; Su, X. Immunomodulatory TGF-β Signaling in Hepatocellular Carcinoma. Trends Mol. Med. 2019, 25, 1010–1023. [Google Scholar] [CrossRef]
- Moon, H.; Ro, S.W. Mapk/Erk Signaling Pathway in Hepatocellular Carcinoma. Cancers 2021, 13, 3026. [Google Scholar] [CrossRef]
- Zhou, Q.; Lui, V.W.Y.; Yeo, W. Targeting the PI3K/Akt/MTOR Pathway in Hepatocellular Carcinoma. Future Oncol. 2011, 7, 1149–1167. [Google Scholar] [CrossRef]
- Mroweh, M.; Roth, G.; Decaens, T.; Marche, P.N.; Lerat, H.; Jílková, Z.M. Targeting AKT in Hepatocellular Carcinoma and Its Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 1794. [Google Scholar] [CrossRef]
- Carbajo-Pescador, S.; Mauriz, J.L.; García-Palomo, A.; González-Gallego, J. FoxO Proteins: Regulation and Molecular Targets in Liver Cancer. Curr. Med. Chem. 2014, 21, 1231–1246. [Google Scholar] [CrossRef]
- Jang, M.-K.; Kim, H.S.; Chung, Y.-H. Clinical Aspects of Tumor Necrosis Factor-α Signaling in Hepatocellular Carcinoma. Curr. Pharm. Des. 2014, 20, 2799–2808. [Google Scholar] [CrossRef]
- Jing, Y.; Sun, K.; Liu, W.; Sheng, D.; Zhao, S.; Gao, L.; Wei, L. Tumor Necrosis Factor-α Promotes Hepatocellular Carcinogenesis through the Activation of Hepatic Progenitor Cells. Cancer Lett. 2018, 434, 22–32. [Google Scholar] [CrossRef]
- Crespo, M.; Leiva, M.; Sabio, G. Circadian Clock and Liver Cancer. Cancers 2021, 13, 3631. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An Expanded Reference Database of Human and Mouse Transcriptional Regulatory Interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, Functions and Interactions with Proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Jiang, M.P.; Xu, W.X.; Hou, J.C.; Xu, Q.; Wang, D.D.; Tang, J.H. The Emerging Role of the Interactions between Circular RNAs and RNA-Binding Proteins in Common Human Cancers. J. Cancer 2021, 12, 5206–5219. [Google Scholar] [CrossRef]
- Paz, I.; Kosti, I.; Ares, M.J.; Cline, M.; Mandel-Gutfreund, Y. RBPmap: A Web Server for Mapping Binding Sites of RNA-Binding Proteins. Nucleic Acids Res. 2014, 42, W361–W367. [Google Scholar] [CrossRef]
- Ray, D.; Kazan, H.; Cook, K.B.; Weirauch, M.T.; Najafabadi, H.S.; Li, X.; Gueroussov, S.; Albu, M.; Zheng, H.; Yang, A.; et al. A Compendium of RNA-Binding Motifs for Decoding Gene Regulation. Nature 2013, 499, 172. [Google Scholar] [CrossRef]
- Cook, K.B.; Kazan, H.; Zuberi, K.; Morris, Q.; Hughes, T.R. RBPDB: A Database of RNA-Binding Specificities. Nucleic Acids Res. 2011, 39, 301–308. [Google Scholar] [CrossRef]
- Chao, H.M.; Huang, H.X.; Chang, P.H.; Tseng, K.C.; Miyajima, A.; Chern, E. Y-Box Binding Protein-1 Promotes Hepatocellular Carcinoma-Initiating Cell Progression and Tumorigenesis via Wnt/β-Catenin Pathway. Oncotarget 2017, 8, 2604–2616. [Google Scholar] [CrossRef]
- Gutschner, T.; Hämmerle, M.; Pazaitis, N.; Bley, N.; Fiskin, E.; Uckelmann, H.; Heim, A.; Gro, M.; Hofmann, N.; Geffers, R.; et al. Insulin-like Growth Factor 2 MRNA-Binding Protein 1 (IGF2BP1) Is an Important Protumorigenic Factor in Hepatocellular Carcinoma. Hepatology 2014, 59, 1900–1911. [Google Scholar] [CrossRef]
- Hu, J.; Cai, D.; Zhao, Z.; Zhong, G.C.; Gong, J. Suppression of Heterogeneous Nuclear Ribonucleoprotein C Inhibit Hepatocellular Carcinoma Proliferation, Migration, and Invasion via Ras/MAPK Signaling Pathway. Front. Oncol. 2021, 11, 1–16. [Google Scholar] [CrossRef]
- Guo, Y.; Zhao, J.; Bi, J.; Wu, Q.; Wang, X.; Lai, Q. Heterogeneous Nuclear Ribonucleoprotein K (HnRNP K) Is a Tissue Biomarker for Detection of Early Hepatocellular Carcinoma in Patients with Cirrhosis. J. Hematol. Oncol. 2012, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.X.; Jiang, S.S.; Zhang, X.F.; Zhou, Z.Q.; Pan, Q.Z.; Chen, C.L.; Zhao, J.J.; Tang, Y.; Xia, J.C.; Weng, D.S. Protein Kinase CK2α Catalytic Subunit Is Overexpressed and Serves as an Unfavorable Prognostic Marker in Primary Hepatocellular Carcinoma. Oncotarget 2015, 6, 34800–34817. [Google Scholar] [CrossRef]
- Jiang, K.; Dong, C.; Yin, Z.; Li, R.; Mao, J.; Wang, C.; Zhang, J.; Gao, Z.; Liang, R.; Wang, Q.; et al. Exosome-Derived ENO1 Regulates Integrin A6β4 Expression and Promotes Hepatocellular Carcinoma Growth and Metastasis. Cell Death Dis. 2020, 11, 972. [Google Scholar] [CrossRef]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA Biology from an Atlas of Mammalian MRNA-Binding Proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef]
- Huppertz, I.; Perez-Perri, J.I.; Mantas, P.; Sekaran, T.; Schwarzl, T.; Dimitrova-Paternoga, L.; Hennig, J.; Neveu, P.A.; Hentze, M.W. RNA Regulates Glycolysis and Embryonic Stem Cell Differentiation via Enolase 1. bioRxiv 2020, 2020. [Google Scholar] [CrossRef]
- Wang, P.; Sun, G.-B.; Dou, G.-X.; Wang, B.-Q. Long Non-coding RNA B3GALT5-AS1 Contributes to the Progression of Gastric Cancer via Interacting with CSNK2A1. Exp. Ther. Med. 2021, 22, 972. [Google Scholar] [CrossRef]
- Mateu-Regué, À.; Christiansen, J.; Bagger, F.O.; Winther, O.; Hellriegel, C.; Nielsen, F.C. Single MRNP Analysis Reveals That Small Cytoplasmic MRNP Granules Represent MRNA Singletons. Cell Rep. 2019, 29, 736–748. [Google Scholar] [CrossRef]
- Li, D.; Liu, X.; Zhou, J.; Hu, J.; Zhang, D.; Liu, J.; Qiao, Y.; Zhan, Q. Long Noncoding RNA HULC Modulates the Phosphorylation of YB-1 through Serving as a Scaffold of Extracellular Signal–Regulated Kinase and YB-1 to Enhance Hepatocarcinogenesis. Hepatology 2017, 65, 1612–1627. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Y.; Yu, S. Long Noncoding RNA AWPPH Promotes Hepatocellular Carcinoma Progression through YBX1 and Serves as a Prognostic Biomarker. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1805–1816. [Google Scholar] [CrossRef]
- Gandhi, M.; Groß, M.; Holler, J.M.; Coggins, S.A.A.; Patil, N.; Leupold, J.H.; Munschauer, M.; Schenone, M.; Hartigan, C.R.; Allgayer, H.; et al. The LncRNA LincNMR Regulates Nucleotide Metabolism via a YBX1-RRM2 Axis in Cancer. Nat. Commun. 2020, 11, 3214. [Google Scholar] [CrossRef]
- Xu, J.; Ji, L.; Liang, Y.; Wan, Z.; Zheng, W.; Song, X.; Gorshkov, K.; Sun, Q.; Lin, H.; Zheng, X.; et al. CircRNA-SORE Mediates Sorafenib Resistance in Hepatocellular Carcinoma by Stabilizing YBX1. Signal Transduct. Target. Ther. 2020, 5, 298. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Xie, X.L.; Zhou, X.; Wang, Y.J.; Wang, Y.J.; Wang, S.L.; Zhang, J.N.; Jiang, X.Y.; Cui, R.L.; Jiang, H.Q.; et al. Y-Box Binding Protein 1 Augments Sorafenib Resistance via the PI3K/Akt Signaling Pathway in Hepatocellular Carcinoma. World J. Gastroenterol. 2021, 27, 4667–4686. [Google Scholar] [CrossRef] [PubMed]
- Habelhah, H.; Shah, K.; Huang, L.; Ostareck-Lederer, A.; Burlingame, A.L.; Shokat, K.M.; Hentze, M.W.; Ronai, Z. ERK Phosphorylation Drives Cytoplasmic Accumulation of HnRNP-K and Inhibition of MRNA Translation. Nat. Cell Biol. 2001, 3, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Otoshi, T.; Tanaka, T.; Morimoto, K.; Nakatani, T. Cytoplasmic Accumulation of Heterogeneous Nuclear Ribonucleoprotein k Strongly Promotes Tumor Invasion in Renal Cell Carcinoma Cells. PLoS ONE 2015, 10, e0145769. [Google Scholar] [CrossRef]
- Alkrekchi, A.; Wang, W.; Rana, P.S.; Markovic, V.; Sossey-Alaoui, K. A Comprehensive Review of the Functions of YB-1 in Cancer Stemness, Metastasis and Drug Resistance. Cell. Signal. 2021, 85, 110073. [Google Scholar] [CrossRef]
- Chen, Z.; Wei, J.; Li, M.; Zhao, Y. A Circular RNAs Dataset Landscape Reveals Potential Signatures for the Detection and Prognosis of Early-Stage Lung Adenocarcinoma. BMC Cancer 2021, 21, 781. [Google Scholar] [CrossRef]
- Sun, S.; Li, C.; Cui, K.; Liu, B.; Zhou, M.; Cao, Y.; Zhang, J.; Bian, Z.; Fei, B.; Huang, Z. Hsa_circ_0062682 Promotes Serine Metabolism and Tumor Growth in Colorectal Cancer by Regulating the MiR-940/PHGDH Axis. Front. Cell Dev. Biol. 2021, 9, 770006. [Google Scholar] [CrossRef]
- Jin, Y.J.; Byun, S.; Han, S.; Chamberlin, J.; Kim, D.; Kim, M.J.; Lee, Y. Differential Alternative Splicing Regulation among Hepatocellular Carcinoma with Different Risk Factors. BMC Med. Genom. 2019, 12, 175. [Google Scholar] [CrossRef]
- Lee, S.E.; Alcedo, K.P.; Kim, H.J.; Snider, N.T. Alternative Splicing in Hepatocellular Carcinoma. Cell. Mol. Gastroenterol. Hepatol. 2020, 10, 699–712. [Google Scholar] [CrossRef]
- Cokan, K.B.; Urlep, Ž.; Lorbek, G.; Matz-Soja, M.; Skubic, C.; Perše, M.; Jeruc, J.; Juvan, P.; Režen, T.; Rozman, D. Chronic Disruption of the Late Cholesterol Synthesis Leads to Female-Prevalent Liver Cancer. Cancers 2020, 12, 3302. [Google Scholar] [CrossRef]
- Wei, J.; Fang, D. Endoplasmic Reticulum Stress Signaling and the Pathogenesis of Hepatocarcinoma. Int. J. Mol. Sci. 2021, 22, 1799. [Google Scholar] [CrossRef]
- Wu, J.; Qiao, S.; Xiang, Y.; Cui, M.; Yao, X.; Lin, R.; Zhang, X. Endoplasmic Reticulum Stress: Multiple Regulatory Roles in Hepatocellular Carcinoma. Biomed. Pharmacother. 2021, 142, 112005. [Google Scholar] [CrossRef]
- Kent, L.N.; Bae, S.; Tsai, S.Y.; Tang, X.; Srivastava, A.; Koivisto, C.; Martin, C.K.; Ridolfi, E.; Miller, G.C.; Zorko, S.M.; et al. Dosage-Dependent Copy Number Gains in E2f1 and E2f3 Drive Hepatocellular Carcinoma. J. Clin. Investig. 2017, 127, 830–842. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, X.; Lu, Y.; Wang, X.; Zhu, L. Hypoxia Inducible Factor 1α in Hepatocellular Carcinoma with Cirrhosis: Association with Prognosis. Pathol. Res. Pract. 2018, 214, 1987–1992. [Google Scholar] [CrossRef]
- Liu, L.; Ji, P.; Qu, N.; Pu, W.L.; Jiang, D.W.; Liu, W.Y.; Li, Y.Q.; Shi, R.L. The Impact of High Co-Expression of Sp1 and HIF1α on Prognosis of Patients with Hepatocellular Cancer. Oncol. Lett. 2016, 12, 504–512. [Google Scholar] [CrossRef]
- Wilson, C.L.; Jurk, D.; Fullard, N.; Banks, P.; Page, A.; Luli, S.; Elsharkawy, A.M.; Gieling, R.G.; Chakraborty, J.B.; Fox, C.; et al. NFκB1 Is a Suppressor of Neutrophil-Driven Hepatocellular Carcinoma. Nat. Commun. 2015, 6, 6818. [Google Scholar] [CrossRef]
- Liao, L.; Chen, C.; Li, N.; Lin, L. Y-Box Binding Protein-1 Promotes Epithelial-Mesenchymal Transition in Sorafenib-Resistant Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2021, 22, 224. [Google Scholar] [CrossRef]
- Liu, X.; Chen, D.; Chen, H.; Wang, W.; Liu, Y.; Wang, Y.; Duan, C.; Ning, Z.; Guo, X.; Otkur, W.; et al. YB1 Regulates MiR-205/200b-ZEB1 Axis by Inhibiting MicroRNA Maturation in Hepatocellular Carcinoma. Cancer Commun. 2021, 41, 576–595. [Google Scholar] [CrossRef]
- Lasham, A.; Samuel, W.; Cao, H.; Patel, R.; Mehta, R.; Stern, J.L.; Reid, G.; Woolley, A.G.; Miller, L.D.; Black, M.A.; et al. YB-1, the E2F Pathway, and Regulation of Tumor Cell Growth. J. Natl. Cancer Inst. 2012, 104, 133–146. [Google Scholar] [CrossRef]
- Higashi, K.; Inagaki, Y.; Suzuki, N.; Mitsui, S.; Mauviel, A.; Kaneko, H.; Nakatsuka, I. Y-Box-Binding Protein YB-1 Mediates Transcriptional Repression of Human A2(I) Collagen Gene Expression by Interferon-γ. J. Biol. Chem. 2003, 278, 5156–5162. [Google Scholar] [CrossRef] [Green Version]
- El-Naggar, A.M.; Veinotte, C.J.; Cheng, H.; Grunewald, T.G.P.; Negri, G.L.; Somasekharan, S.P.; Corkery, D.P.; Tirode, F.; Mathers, J.; Khan, D.; et al. Translational Activation of HIF1α by YB-1 Promotes Sarcoma Metastasis. Cancer Cell 2015, 27, 682–697. [Google Scholar] [CrossRef]
- Prabhu, L.; Mundade, R.; Wang, B.; Wei, H.; Hartley, A.V.; Martin, M.; McElyea, K.; Temm, C.J.; Sandusky, G.; Liu, Y.; et al. Critical Role of Phosphorylation of Serine 165 of YBX1 on the Activation of NF-ΚB in Colon Cancer. Oncotarget 2015, 6, 29396–29412. [Google Scholar] [CrossRef]
- Martin, M.; Hua, L.; Wang, B.; Wei, H.; Prabhu, L.; Hartley, A.V.; Jiang, G.; Liu, Y.; Lu, T. Novel Serine 176 Phosphorylation of YBX1 Activates NF-ΚB in Colon Cancer. J. Biol. Chem. 2017, 292, 3433–3444. [Google Scholar] [CrossRef]
- Homer, C.; Knight, D.A.; Hananeia, L.; Sheard, P.; Risk, J.; Lasham, A.; Royds, J.A.; Braithwaite, A.W. Y-Box Factor YB1 Controls P53 Apoptotic Function. Oncogene 2005, 24, 8314–8325. [Google Scholar] [CrossRef]
- Evans, M.K.; Matsui, Y.; Xu, B.; Willis, C.; Loome, J.; Milburn, L.; Fan, Y.; Pagala, V.; Peng, J.C. Ybx1 Fine-Tunes PRC2 Activities to Control Embryonic Brain Development. Nat. Commun. 2020, 11, 4060. [Google Scholar] [CrossRef]
- Ha, B.; Lee, E.B.; Cui, J.; Kim, Y.; Jang, H.H. YB-1 Overexpression Promotes a TGF-Β1-Induced Epithelial-Mesenchymal Transition via Akt Activation. Biochem. Biophys. Res. Commun. 2015, 458, 347–351. [Google Scholar] [CrossRef]
- Jayavelu, A.K.; Schnöder, T.M.; Perner, F.; Herzog, C.; Meiler, A.; Krishnamoorthy, G.; Huber, N.; Mohr, J.; Edelmann-Stephan, B.; Austin, R.; et al. Splicing Factor YBX1 Mediates Persistence of JAK2-Mutated Neoplasms. Nature 2020, 588, 157–163. [Google Scholar] [CrossRef]
- Jürchott, K.; Kuban, R.-J.; Krech, T.; Blüthgen, N.; Stein, U.; Walther, W.; Friese, C.; Kiełbasa, S.M.; Ungethüm, U.; Lund, P.; et al. Identification of Y-Box Binding Protein 1 As a Core Regulator of MEK/ERK Pathway-Dependent Gene Signatures in Colorectal Cancer Cells. PLOS Genet. 2010, 6, e1001231. [Google Scholar] [CrossRef]
- Delicato, A.; Montuori, E.; Angrisano, T.; Pollice, A.; Calabrò, V. Yb-1 Oncoprotein Controls Pi3k/Akt Pathway by Reducing Pten Protein Level. Genes 2021, 12, 1551. [Google Scholar] [CrossRef]
- Shah, A.; Plaza-Sirvent, C.; Weinert, S.; Buchbinder, J.H.; Lavrik, I.N.; Mertens, P.R.; Schmitz, I.; Lindquist, J.A. YB-1 Mediates TNF-Induced Pro-Survival Signaling by Regulating NF-ΚB Activation. Cancers 2020, 12, 2188. [Google Scholar] [CrossRef]
- Hessman, C.L.; Hildebrandt, J.; Shah, A.; Brandt, S.; Bock, A.; Frye, B.C.; Raffetseder, U.; Geffers, R.; Brunner-Weinzierl, M.C.; Isermann, B.; et al. YB-1 Interferes with Tnfα–Tnfr Binding and Modulates Progranulin-Mediated Inhibition of TNFα Signaling. Int. J. Mol. Sci. 2020, 21, 7076. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Kose, S.; Kuwahara, I.; Yoshimura, M.; Imamoto, N.; Yoshida, M. Y-Box Protein-Associated Acidic Protein (YBAP1/C1QBP) Affects the Localization and Cytoplasmic Functions of YB-1. Sci. Rep. 2018, 8, 6198. [Google Scholar] [CrossRef] [PubMed]
- Kwon, E.; Todorova, K.; Wang, J.; Horos, R.; Lee, K.K.; Neel, V.A.; Negri, G.L.; Sorensen, P.H.; Lee, S.W.; Hentze, M.W.; et al. The RNA-Binding Protein YBX1 Regulates Epidermal Progenitors at a Posttranscriptional Level. Nat. Commun. 2018, 9, 1734. [Google Scholar] [CrossRef]
- Kosnopfel, C.; Sinnberg, T.; Sauer, B.; Busch, C.; Niessner, H.; Schmitt, A.; Forchhammer, S.; Grimmel, C.; Mertens, P.R.; Hailfinger, S.; et al. YB-1 Expression and Phosphorylation Regulate Tumorigenicity and Invasiveness in Melanoma by Influencing EMT. Mol. Cancer Res. 2018, 16, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, J.; Ziol, M.; Paradis, V.; Zucman-Rossi, J. Molecular and Histological Correlations in Liver Cancer. J. Hepatol. 2019, 71, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhou, Z.; Hu, A.; Pontede Albuquerque, C.; Zhou, Y.; Hong, L.; Sierecki, E.; Ajiro, M.; Kruhlak, M.; Harris, C.; et al. Both Decreased and Increased SRPK1 Levels Promote Cancer by Interfering with PHLPP-Mediated Dephosphorylation of Akt. Mol. Cell 2014, 54, 378–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razpotnik, R.; Vidmar, R.; Fonović, M.; Rozman, D.; Režen, T. Circular RNA hsa_circ_0062682 Binds to YBX1 and Promotes Oncogenesis in Hepatocellular Carcinoma. Cancers 2022, 14, 4524. https://doi.org/10.3390/cancers14184524
Razpotnik R, Vidmar R, Fonović M, Rozman D, Režen T. Circular RNA hsa_circ_0062682 Binds to YBX1 and Promotes Oncogenesis in Hepatocellular Carcinoma. Cancers. 2022; 14(18):4524. https://doi.org/10.3390/cancers14184524
Chicago/Turabian StyleRazpotnik, Rok, Robert Vidmar, Marko Fonović, Damjana Rozman, and Tadeja Režen. 2022. "Circular RNA hsa_circ_0062682 Binds to YBX1 and Promotes Oncogenesis in Hepatocellular Carcinoma" Cancers 14, no. 18: 4524. https://doi.org/10.3390/cancers14184524
APA StyleRazpotnik, R., Vidmar, R., Fonović, M., Rozman, D., & Režen, T. (2022). Circular RNA hsa_circ_0062682 Binds to YBX1 and Promotes Oncogenesis in Hepatocellular Carcinoma. Cancers, 14(18), 4524. https://doi.org/10.3390/cancers14184524