Cytology Smears: An Enhanced Alternative Method for Colorectal Cancer pN Stage—A Multicentre Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Inclusion Criteria
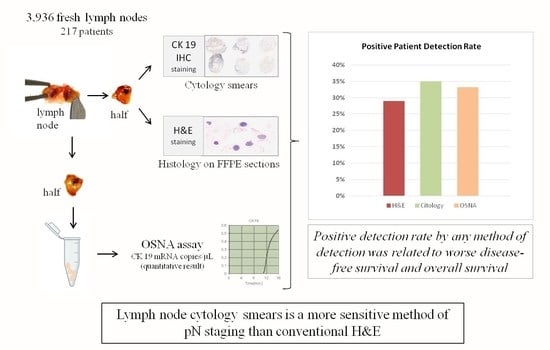
2.2. Fresh Lymph Node Dissection and LN Analysis
2.3. One-Step Nucleic Acid Amplification (OSNA) Assay
2.4. CK19 Immunochemistry
2.5. Lymph Node Pathology Reporting and pN Staging
2.6. Statistical Analysis
2.7. Study Endpoints and Survival Analysis
3. Results
3.1. Clinical and Pathological Characteristics
3.2. Patient Positive Detection Rate Is Higher with Cytology Smears and OSNA Than with H&E
3.3. pN Upstaging with LN Cytology Smears with Respect to H&E and High Diagnostic Efficacy of Cytology Smears to Discriminate LN-Positive Patients
3.4. The Total Tumour Load (TTL) Increases with the pN Stage
3.5. Patients with Positive LNs Exhibit Worse Survival Outcome as Determined by Any of the Three Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gunderson, L.L.; Jessup, J.M.; Sargent, D.J.; Greene, F.L.; Stewart, A.K. Revised TN categorization for colon cancer based on national survival outcomes data. J. Clin. Oncol. 2010, 28, 264–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NCCN. Colon Cancer. In Encyclopedia of Genetics, Genomics, Proteomics and Informatics; Springer: Dordrecht, The Nertherland, 2018; p. 392. [Google Scholar] [CrossRef]
- Williams, J.L.; Chan, C.K.; Toste, P.A.; Elliott, I.A.; Vasquez, C.R.; Sunjaya, D.B.; Swanson, E.A.; Koo, J.; Hines, O.J.; Reber, H.A.; et al. Association of histopathologic phenotype of periampullary adenocarcinomas with survival. JAMA Surg. 2017, 152, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Kubisch, C.H.; Crispin, A.; Mansmann, U.; Göke, B.; Kolligs, F.T. Screening for Colorectal Cancer is Associated with Lower Disease Stage: A Population-Based Study. Clin. Gastroenterol. Hepatol. 2016. [Google Scholar] [CrossRef]
- Quintero, E.; Castells, A.; Bujanda, L.; Cubiella, J.; Salas, D.; Lanas, Á.; Andreu, M.; Carballo, F.; Morillas, J.D.; Hernández, C.; et al. Colonoscopy versus Fecal Immunochemical Testing in Colorectal-Cancer Screening. N. Engl. J. Med. 2012, 366, 697–706. [Google Scholar] [CrossRef] [Green Version]
- Schreuders, E.H.; Ruco, A.; Rabeneck, L.; Schoen, R.E.; Sung, J.J.Y.; Young, G.P.; Kuipers, E.J. Colorectal cancer screening: A global overview of existing programmes. Gut 2015, 64, 1637–1649. [Google Scholar] [CrossRef] [PubMed]
- Castells, A.; Quintero, E.; Álvarez, C.; Bujanda, L.; Cubiella, J.; Salas, D.; Lanas, A.; Carballo, F.; Morillas, J.D.; Hernández, C.; et al. Rate of detection of advanced neoplasms in proximal colon by simulated sigmoidoscopy vs. fecal immunochemical tests. Clin. Gastroenterol. Hepatol. 2014, 12, 1708–1716.e4. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, D.; Atkin, W.; Lenz, H.-J.; Lynch, H.T.; Minsky, B.; Nordlinger, B.; Starling, N. Colorectal cancer. Lancet 2010, 375, 1030–1047. [Google Scholar] [CrossRef] [PubMed]
- Rahbari, N.N.; Bork, U.; Motschall, E.; Thorlund, K.; Büchler, M.W.; Koch, M.; Weitz, J. Molecular detection of tumor cells in regional lymph nodes is associated with disease recurrence and poor survival in node-negative colorectal cancer: A systematic review and meta-analysis. J. Clin. Oncol. 2012, 30, 60–70. [Google Scholar] [CrossRef]
- Compton, C.; Fenoglio-Preiser, C.M.; Pettigrew, N.; Fielding, L.P. American Joint Committee on Cancer prognostic factors consensus conference: Colorectal working group. Cancer 2000, 88, 1739–1757. [Google Scholar] [CrossRef]
- Washington, M.K.; Berlin, J.; Branton, P.; Burgart, L.J.; Carter, D.K.; Fitzgibbons, P.L.; Halling, K.; Frankel, W.; Jessup, J.; Kakar, S.; et al. Protocol for the Examination of Specimens from Patients with Primary Carcinoma of the Colon and Rectum. College of American Pathologists. Arch. Pathol. Lab. Med. 2009, 133, 1539–1551. [Google Scholar] [CrossRef]
- Itabashi, M.; Yamamoto, H.; Tomita, N.; Inomata, M.; Murata, K.; Hayashi, S.; Miyake, Y.; Igarashi, S.; Kato, T.; Noura, S.; et al. Lymph Node Positivity in One-Step Nucleic Acid Amplification is a Prognostic Factor for Postoperative Cancer Recurrence in Patients with Stage II Colorectal Cancer: A Prospective, Multicenter Study. Ann. Surg. Oncol. 2020, 27, 1077–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iddings, D.; Ahmad, A.; Elashoff, D.; Bilchik, A. The prognostic effect of micrometastases in previously staged lymph node negative (N0) colorectal carcinoma: A meta-analysis. Ann. Surg. Oncol. 2006, 13, 1386–1392. [Google Scholar] [CrossRef] [PubMed]
- Aldecoa, I.; Atares, B.; Tarragona, J.; Bernet, L.; Sardon, J.D.; Pereda, T.; Villar, C.; Mendez, M.C.; Gonzalez-Obeso, E.; Elorriaga, K.; et al. Molecularly determined total tumour load in lymph nodes of stage I-II colon cancer patients correlates with high-risk factors. A multicentre prospective study. Virchows Arch. 2016, 469, 385–394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archilla, I.; Díaz-Mercedes, S.; Aguirre, J.J.; Tarragona, J.; Machado, I.; Rodrigo, M.T.; Lopez-Prades, S.; Gorostiaga, I.; Landolfi, S.; Alén, B.O.; et al. Lymph Node Tumor Burden Correlates With Tumor Budding and Poorly Differentiated Clusters: A New Prognostic Factor in Colorectal Carcinoma? Clin. Transl. Gastroenterol. 2021, 12, e00303. [Google Scholar] [CrossRef]
- Espinosa-Bravo, M.; Sansano, I.; Pérez-Hoyos, S.; Ramos, M.; Sancho, M.; Xercavins, J.; Rubio, I.T.; Peg, V. Prediction of non-sentinel lymph node metastasis in early breast cancer by assessing total tumoral load in the sentinel lymph node by molecular assay. Eur. J. Surg. Oncol. 2013, 39, 766–773. [Google Scholar] [CrossRef]
- Rubio, I.T.; Espinosa-Bravo, M.; Rodrigo, M.; Amparo Viguri Diaz, M.; Hardisson, D.; Sagasta, A.; Dueñas, B.; Peg, V. Nomogram including the total tumoral load in the sentinel nodes assessed by one-step nucleic acid amplification as a new factor for predicting nonsentinel lymph node metastasis in breast cancer patients. Breast Cancer Res. Treat. 2014, 147, 371–380. [Google Scholar] [CrossRef]
- Cordoba, O.; Perez-Ceresuela, F.; Espinosa-Bravo, M.; Cortadellas, T.; Esgueva, A.; Rodriguez-Revuelto, R.; Peg, V.; Reyes, V.; Xercavins, J.; Rubio, I.T. Detection of sentinel lymph node in breast cancer recurrence may change adjuvant treatment decision in patients with breast cancer recurrence and previous axillary surgery. Breast 2014, 23, 460–465. [Google Scholar] [CrossRef]
- Peg, V.; Espinosa-Bravo, M.; Vieites, B.; Vilardell, F.; Antúnez, J.R.; de Salas, M.S.; Delgado-Sánchez, J.J.; Pinto, W.; Gozalbo, F.; Petit, A.; et al. Intraoperative molecular analysis of total tumor load in sentinel lymph node: A new predictor of axillary status in early breast cancer patients. Breast Cancer Res. Treat. 2013, 139, 87–93. [Google Scholar] [CrossRef]
- Espinosa-Bravo, M.; Navarro-Cecilia, J.; Ramos Boyero, M.; Diaz-Botero, S.; Dueñas Rodríguez, B.; Luque López, C.; Ramos Grande, T.; Ruano Perez, R.; Peg, V.; Rubio, I.T. Intraoperative assessment of sentinel lymph node by one-step nucleic acid amplification in breast cancer patients after neoadjuvant treatment reduces the need for a second surgery for axillary lymph node dissection. Breast 2017, 31, 40–45. [Google Scholar] [CrossRef]
- Peg, V.; Sansano, I.; Vieites, B.; Bernet, L.; Cano, R.; Córdoba, A.; Sancho, M.; Martín, M.D.; Vilardell, F.; Cazorla, A.; et al. Role of total tumour load of sentinel lymph node on survival in early breast cancer patients. Breast 2017, 33, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, H.; Sekimoto, M.; Oya, M.; Yamamoto, N.; Konishi, F.; Sasaki, J.; Yamada, S.; Taniyama, K.; Tominaga, H.; Tsujimoto, M.; et al. OSNA-based novel molecular testing for lymph node metastases in colorectal cancer patients: Results from a multicenter clinical performance study in Japan. Ann. Surg. Oncol. 2011, 18, 1891–1898. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Daito, M.; Hiyama, K.; Ding, J.; Nakabayashi, K.; Otomo, Y.; Tsujimoto, M.; Matsuura, N.; Kato, Y. An optimal mRNA marker for OSNA (One-step nucleic acid amplification) based lymph node metastasis detection in colorectal cancer patients. Jpn. J. Clin. Oncol. 2013, 43, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Tomita, N.; Inomata, M.; Furuhata, T.; Miyake, Y.; Noura, S.; Kato, T.; Murata, K.; Hayashi, S.; Igarashi, S.; et al. OSNA-Assisted Molecular Staging in Colorectal Cancer: A Prospective Multicenter Trial in Japan. Ann. Surg. Oncol. 2016, 23, 391–396. [Google Scholar] [CrossRef] [PubMed]
- McGovern, J.; Fuller, C.; Burris, K. Anal cancer screening and prevention: A review for dermatologists. J. Eur. Acad. Dermatology Venereol. 2021, 35, 1622–1627. [Google Scholar] [CrossRef]
- Kyrgiou, M.; Arbyn, M.; Bergeron, C.; Bosch, F.X.; Dillner, J.; Jit, M.; Kim, J.; Poljak, M.; Nieminen, P.; Sasieni, P.; et al. Cervical screening: ESGO-EFC position paper of the European Society of Gynaecologic Oncology (ESGO) and the European Federation of Colposcopy (EFC). Br. J. Cancer 2020, 123, 510–517. [Google Scholar] [CrossRef]
- Al-Abbadi, M.A. Basics of cytology. Avicenna J. Med. 2011, 1, 18–28. [Google Scholar] [CrossRef]
- Harnish, B.; Nidhi, V.; Neena, D. Usefulness of touch Imprint Cytology in Cancer diagnosis: A study of 119 cases. Int. Res. J. Med. Sci. Int. Res. J. Med. Sci 2014, 2, 2320–7353. [Google Scholar]
- Iacuzzo, C.; Troian, M.; Bonadio, L.; Bonazza, D.; Dobrinja, C.; Bellio, G.; Scomersi, S.; Giudici, F.; Zanconati, F.; Bortul, M. Evaluation of Sentinel Lymph Node Intraoperative Touch Imprint Cytology in Breast Cancer Surgery. J. Mol. Biomark. Diagn. 2017, 8, 6–10. [Google Scholar] [CrossRef] [Green Version]
- Rakislova, N.; Montironi, C.; Aldecoa, I.; Fernandez, E.; Bombi, J.A.; Jimeno, M.; Balaguer, F.; Pellise, M.; Castells, A.; Cuatrecasas, M. Lymph node pooling: A feasible and efficient method of lymph node molecular staging in colorectal carcinoma. J. Transl. Med. 2017, 15, 14. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef]
- Tsujimoto, M.; Nakabayashi, K.; Yoshidome, K.; Kaneko, T.; Iwase, T.; Akiyama, F.; Kato, Y.; Tsuda, H.; Ueda, S.; Sato, K.; et al. One-step nucleic acid amplification for intraoperative detection of lymph node metastasis in breast cancer patients. Clin. Cancer Res. 2007, 13, 4807–4816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croner, R.S.; Geppert, C.-I.; Bader, F.G.; Nitsche, U.; Späth, C.; Rosenberg, R.; Zettl, A.; Matias-Guiu, X.; Tarragona, J.; Güller, U.; et al. Molecular staging of lymph node-negative colon carcinomas by one-step nucleic acid amplification (OSNA) results in upstaging of a quarter of patients in a prospective, European, multicentre study. Br. J. Cancer 2014, 110, 2544–2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Påhlman, L.A.; Hohenberger, W.M.; Matzel, K.; Sugihara, K.; Quirke, P.; Glimelius, B. Should the Benefit of Adjuvant Chemotherapy in Colon Cancer Be Re-Evaluated? J. Clin. Oncol. 2016, 34, 1297–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aldecoa, I.; Montironi, C.; Planell, N.; Pellise, M.; Fernandez-Esparrach, G.; Gines, A.; Delgado, S.; Momblan, D.; Moreira, L.; Lopez-Ceron, M.; et al. Endoscopic tattooing of early colon carcinoma enhances detection of lymph nodes most prone to harbor tumor burden. Surg. Endosc. Other Interv. Tech. 2016, 31, 723–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, H.; Murata, K.; Fukunaga, M.; Ohnishi, T.; Noura, S.; Miyake, Y.; Kato, T.; Ohtsuka, M.; Nakamura, Y.; Takemasa, I.; et al. Micrometastasis Volume in Lymph Nodes Determines Disease Recurrence Rate of Stage II Colorectal Cancer: A Prospective Multicenter Trial. Clin. Cancer Res. 2016, 22, 3201–3208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Güller, U.; Zettl, A.; Worni, M.; Langer, I.; Cabalzar-Wondberg, D.; Viehl, C.T.; Demartines, N.; Zuber, M. Molecular investigation of lymph nodes in colon cancer patients using one-step nucleic acid amplification (OSNA): A new road to better staging? Cancer 2012, 118, 6039–6045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croner, R.S.; Schellerer, V.; Demund, H.; Schildberg, C.; Papadopulos, T.; Naschberger, E.; Stürzl, M.; Matzel, K.E.; Hohenberger, W.; Schlabrakowski, A. One step nucleic acid amplification (OSNA)—A new method for lymph node staging in colorectal carcinomas. J. Transl. Med. 2010, 8, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanebo, H.J.; LeGolvan, M.; Paty, P.B.; Saha, S.; Zuber, M.; D’Angelica, M.I.; Kemeny, N.E. Meeting the biologic challenge of colorectal metastases. Clin. Exp. Metastasis 2012, 29, 821–839. [Google Scholar] [CrossRef]
- Hiyoshi, Y.; Akiyoshi, T.; Fukunaga, Y. The advantage of one-step nucleic acid amplification for the diagnosis of lymph node metastasis in colorectal cancer patients. Ann. Gastroenterol. Surg. 2020, 5, 60–66. [Google Scholar] [CrossRef]
- Sansano, I.; Vieites, B.; Sancho de Salas, M.; García, C.; Amendoeira, I.; Bernet, L.; Pérez-García, J.M.; Espinosa-Bravo, M.; Rubio, I.T.; Ramón y Cajal, S.; et al. Axillary staging based on molecular analysis: Results of the B-CLOSER-II study. Pathol.—Res. Pract. 2020, 216, 153197. [Google Scholar] [CrossRef]
- Vieites, B.; López-García, M.; Martín-Salvago, M.D.; Ramirez-Tortosa, C.L.; Rezola, R.; Sancho, M.; López-Vilaró, L.; Villardell, F.; Burgués, O.; Fernández-Rodriguez, B.; et al. Predictive and prognostic value of total tumor load in sentinel lymph nodes in breast cancer patients after neoadjuvant treatment using one-step nucleic acid amplification: The NEOVATTL study. Clin. Transl. Oncol. 2021, 23, 1377–1385. [Google Scholar] [CrossRef] [PubMed]
- Cipolla, C.; Graceffa, G.; Cabibi, D.; Gangi, G.; Latteri, M.; Valerio, M.R.; Vieni, S. Current role of intraoperative frozen section examination of sentinel lymph node in early breast cancer. Anticancer Res. 2020, 40, 1711–1717. [Google Scholar] [CrossRef] [PubMed]





| Variable | Strata | Patients (N = 217) | |
|---|---|---|---|
| n % | |||
| Age | Median, years (range) | 71 (39–92) | |
| Sex | Male | 129 | 59.4% |
| Female | 88 | 40.6% | |
| Histological grade | High | 87 | 40.1% |
| Low | 130 | 59.9% | |
| pT stage | T1 | 42 | 19.4% |
| T2 | 61 | 28.1% | |
| T3 | 97 | 44.7% | |
| T4 | 17 | 7.8% | |
| pN stage by H&E | N0 | 154 | 71% |
| N1 | 46 | 21.2% | |
| N2 | 17 | 7.8% | |
| pN stage by cytology | N0 | 141 | 65% |
| N1 | 55 | 25.3% | |
| N2 | 21 | 9.7% | |
| Total tumor load (CK19 mRNA copies/uL) | Median (range) | 19,225.16 (0–1,600,000) | |
| <250 | 145 | 66.8% | |
| 250 to 6000 | 15 | 6.9% | |
| >6000 | 57 | 26.3% | |
| Number of Lymph Nodes (Total: 4310) | Fresh LN | 3936 | 91.32% |
| Post-formalin fixation LN | 374 | 8.68% | |
| Lymph nodes (LNs) analysed per patient | Total (median, range) | 18 (4–62) | |
| Fresh (median, range) | 17 (3–62) | ||
| Post-fixation formol (median, range) | 0 (0–27) | ||
| Recurrence | Yes | 33 | 15.2% |
| No | 184 | 84.8% | |
| Adjuvant chemotherapy | Yes | 55 | 25.5% |
| No | 161 | 74.5% | |
| Lymphovascular invasion | Yes | 73 | 66.4% |
| No | 144 | 33.6% | |
| Perineural invasion | Yes | 45 | 20.7% |
| No | 172 | 79.3% | |
| Tumour budding | Bd1 | 120 | 55.3% |
| Bd2 | 53 | 24.4% | |
| Bd3 | 44 | 20.3% | |
| H&E | Cytology | OSNA | ||
|---|---|---|---|---|
| Proportion of Patients/Detection Rate | Number (%) | 217 (100%) | 217 (100%) | 217 (100%) |
| Positive LNs | 63 (29%) | 76 (35%) | 72 (33.2%) | |
| Negative LNs | 154 (71%) | 141 (65%) | 145 (66.8%) | |
| Total number of LNs analysed | Number (%) | 3936 (100%) | 3936 (100%) | 3926 (100%) |
| Positive | 210 (5.3%) | 249 (6.3%) | - | |
| Negative | 3726 (94.7%) | 3687 (93.7%) | - | |
| Average number of LNs analysed per patient | Mean (range) | 18.1 (3–62) | 18.1 (3–62) | - |
| Positive | 0.97 (0–17) | 1.15 (0–17) | - | |
| Negative | 17.2 (1–62) | 17 (3–62) | - |
| pN Staging by H&E | pN Staging by Cytology | OSNA | ||||||
|---|---|---|---|---|---|---|---|---|
| pN0 | pN1a+b | pN2a+b | pN0 | pN1a+b | pN2a+b | 0 to <250 | 250 to <6000 | ≥6000 |
| 154 (71%) | 46 (21.2%) | 17 (7.8%) | 141 (65%) | 55 (25.3%) | 21 (9.7%) | 145 (66.8%) | 57 (26.3%) | 15 (6.9%) |
| Negative | Positive | Negative | Positive | Negative (<250) | Positive (≥250) | |||
| 154 (71%) | 63 (29%) | 141 (65%) | 76 (35%) | 145 (66.8%) | 72 (33.2%) | |||
| Concordant cases, number (%) | 200 (92.2%) | 192 (88.5%) | ||||||
| Discordant cases, number (%) | 17 (7.8%) | 25 (11.5%) | ||||||
| Kappa Index | 82.1% | 73.2% | ||||||
| McNemar’s p-value | 0.004 | 0.11 | ||||||
| Sensitivity (%) | 96.8% | 87.3% | ||||||
| Specificity (%) | 90.3% | 89% | ||||||
| Positive Predictive Value (%) | 80.3% | 76.4% | ||||||
| Negative Predictive Value (%) | 98.6% | 94.5% | ||||||
| Number of cases | Cytology-negative | Cytology-positive | OSNA-negative | OSNA-positive | ||||
| H&E-negative | 139 | 15 | 137 | 17 | ||||
| H&E-positive | 2 | 61 | 8 | 55 | ||||
| Variable | Strata | Disease-Free Survival (RFS) | Overall Survival (OS) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate Analysis | Multivariable Analysis | Univariate Analysis | Multivariable Analysis | ||||||
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | HR (95% CI) | p-Value | HR (95% CI) | p-Value | ||
| pN stage H&E | N1/N2 vs. N0 | 4.85 (2.69–8.74) | <0.0001 | 4.27 (2.14–8.49) | <0.0001 | 2.70 (1.34–5.47) | 0.004 | 2.25 (1–5.08) | 0.049 |
| pN stage cytology | N1/N2 vs. N0 | 4.50 (2.44–8.32) | <0.0001 | 3.87 (1.95–7.69) | 0.0001 | 2.54 (1.25–5.19) | 0.008 | 2.20 (0.98–4.94) | 0.057 |
| TTL | Positive vs. Negative | 3.80 (2.11–6.85) | <0.0001 | 2.90 (1.53–5.50) | 0.001 | 2.51 (1.24–5.09) | 0.02 | 1.97 (0.91–4.26) | 0.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diaz-Mercedes, S.; Archilla, I.; Lahoz, S.; Rodrigo-Calvo, M.T.; Lopez-Prades, S.; Tarragona, J.; Landolfi, S.; Concha, A.; Machado, I.; Maurel, J.; et al. Cytology Smears: An Enhanced Alternative Method for Colorectal Cancer pN Stage—A Multicentre Study. Cancers 2022, 14, 6072. https://doi.org/10.3390/cancers14246072
Diaz-Mercedes S, Archilla I, Lahoz S, Rodrigo-Calvo MT, Lopez-Prades S, Tarragona J, Landolfi S, Concha A, Machado I, Maurel J, et al. Cytology Smears: An Enhanced Alternative Method for Colorectal Cancer pN Stage—A Multicentre Study. Cancers. 2022; 14(24):6072. https://doi.org/10.3390/cancers14246072
Chicago/Turabian StyleDiaz-Mercedes, Sherley, Ivan Archilla, Sara Lahoz, Maria Teresa Rodrigo-Calvo, Sandra Lopez-Prades, Jordi Tarragona, Stefania Landolfi, Angel Concha, Isidro Machado, Joan Maurel, and et al. 2022. "Cytology Smears: An Enhanced Alternative Method for Colorectal Cancer pN Stage—A Multicentre Study" Cancers 14, no. 24: 6072. https://doi.org/10.3390/cancers14246072
APA StyleDiaz-Mercedes, S., Archilla, I., Lahoz, S., Rodrigo-Calvo, M. T., Lopez-Prades, S., Tarragona, J., Landolfi, S., Concha, A., Machado, I., Maurel, J., Chic, N., Castells, A., Balaguer, F., Camps, J., & Cuatrecasas, M. (2022). Cytology Smears: An Enhanced Alternative Method for Colorectal Cancer pN Stage—A Multicentre Study. Cancers, 14(24), 6072. https://doi.org/10.3390/cancers14246072