Circulating Proteins Associated with Response and Resistance to Neoadjuvant Chemotherapy in HER2-Positive Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
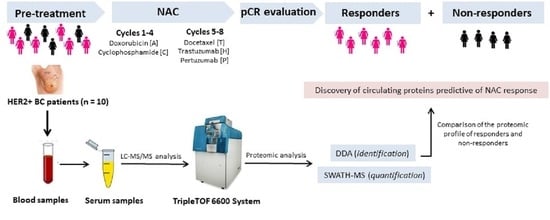
2. Materials and Methods
2.1. Patient Study Group
2.2. Collection and Storage of Blood Serum
2.3. Chemicals and Reagents
2.4. Synthesis of Inorganic Nanoparticles
2.5. Instrumentation
2.6. Depletion of Multiple High-Abundance Proteins in Serum Samples
2.7. Isolation, Fractionation and Digestion of Low-Abundance Proteins
2.8. Qualitative Proteomic Analysis by Mass Spectrometry (LC-MS/MS): Identification by Data-dependent Acquisition (DDA)
2.9. Quantitative Proteomic Analysis by Sequential Window Acquisition of All Theoretical Mass Spectrometry (SWATH-MS)
2.10. Protein Functional Interaction Network Analysis
2.11. Statistical Analysis
2.12. Development of the Classifiers
3. Results
3.1. Clinicopathological Features of Patients
3.2. Proteomic Discovery Using the DDA Approach
- (a)
- complements: complement C1q subcomponent subunit B (C1QB), complement C1q subcomponent subunit C (C1QC), complement C2 (C2), complement C3 (C3), complement C4-B (C4B), complement factor B (CFB);
- (b)
- serine protease related proteins: antithrombin-III (SERPINC1), alpha-2-antiplasmin (SERPINF2), plasma protease C1 inhibitor (SERPING1);
- (c)
- vitamin K-dependent proteins: vitamin K-dependent protein S (PROS1), and
- (d)
- glycoproteins: vitronectin (VTN),
- (e)
- other groups: alpha-2-macroglobulin (A2M), clusterin (CLU), and kininogen-1 (KNG1).
3.3. Differential Protein Expression
3.4. Comparison of the Serum Proteomic Profile Common to the Three Methods Obtained by Shotgun (DDA Analysis) and SWATH-MS in HER2-Positive BC Patients before NAC
3.5. In Silico Validation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thompson, A.M.; Moulder-Thompson, S.L. Neoadjuvant treatment of breast cancer. Ann. Oncol. 2012, 23 (Suppl. S10), x231–x236. [Google Scholar] [CrossRef] [PubMed]
- Buzdar, A.U.; Valero, V.; Theriault, R.; Frye, D.; Green, M.; Booser, D.; Guerra, L.; Sahin, A.; Ames, F.; Smith, T.; et al. Pathological complete response (pCR) to chemotherapy is related to hormone receptor status. Breast Cancer Res. Treat. 2003, 82 (Suppl. S1), 302A. [Google Scholar]
- Petrelli, F.; Borgonovo, K.; Cabiddu, M.; Ghilardi, M.; Barni, S. Neoadjuvant chemotherapy and concomitant trastuzumab in breast cancer: A pooled analysis of two randomized trials. AntiCancer Drugs 2011, 22, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Piccart-Gebhart, M.; Holmes, A.P.; de Azambuja, E.; Di Cosimo, S.; Swaby, R.; Untch, M.; Jackisch, C.; Lang, I.; Smith, I.; Boyle, F.; et al. The association between event-free survival and pathological complete response to neoadjuvant lapatinib, trastuzumab or their combination in HER2-positive breast cancer. Survival follow-up analysis of the NeoALTTO study (BIG 1-06). Cancer Res. 2013, 73, S1-01. [Google Scholar]
- Gianni, L.; Pienkowski, T.; Im, Y.H.; Roman, L.; Tseng, L.-M.; Liu, M.-C.; Lluch, A.; Staroslawska, E.; Haba-Rodriguez, J.; Im, S.-A.; et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): A randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012, 13, 25–32. [Google Scholar] [CrossRef]
- Schneeweiss, A.; Chia, S.; Hickish, T.; Harvey, V.; Eniu, A.; Hegg, R.; Tausch, C.; Seo, J.H.; Tsai, Y.-F.; Ratnayake, J.; et al. Pertuzumab plus trastuzumab in combination with standard neoadjuvant anthracycline-containing and anthracycline-free chemotherapy regimens in patients with HER2-positive early breast cancer: A randomized phase II cardiac safety study (TRYPHAENA). Ann. Oncol. 2013, 24, 2278–2284. [Google Scholar] [CrossRef]
- Loibl, S.; Jackisch, C.; Schneeweiss, A.; Schmatloch, S.; Aktas, B.; Denkert, C.; Wiebringhaus, H.; Kümmel, S.; Warm, M.; Paepke, S.; et al. Dual HER2-blockade with pertuzumab and trastuzumab in HER2-positive early breast cancer: A subanalysis of data from the randomized phase III GeparSepto trial. Ann. Oncol. 2017, 28, 497–504. [Google Scholar] [CrossRef]
- Hurvitz, S.A.; Martin, M.; Symmans, W.F.; Jung, K.H.; Huang, C.-S.; Thompson, A.M.; Harbeck, N.; Valero, V.; Stroyakovskiy, D.; Wildiers, H.; et al. Pathologic complete response (pCR) rates after neoadjuvant trastuzumab emtansine (T-DM1 [K]) + pertuzumab (P) vs docetaxel + carboplatin + trastuzumab + P (TCHP) treatment in patients with HER2-positive (HER2+) early breast cancer (EBC) (KRISTINE). J. Clin. Oncol. 2016, 34 (Suppl. S15), 500A. [Google Scholar] [CrossRef]
- Beitsch, P.; Whitworth, P.; Baron, P.; Rotkis, M.C.; Mislowsky, A.M.; Richards, P.D.; Murray, M.K.; Pellicane, J.V.; Dul, C.L.; Nash, C.H.; et al. Pertuzumab/trastuzumab/CT versus trastuzumab/CT therapy for HER2+ breast cancer: Results from the prospective Neoadjuvant Breast Registry Symphony Trial (NBRST). Ann. Surg. Oncol. 2017, 24, 2539–2546. [Google Scholar] [CrossRef]
- Untch, M.; Loibl, S.; Bischoff, J.; Eidtmann, H.; Kaufmann, M.; Blohmer, J.-U.; Hilfrich, J.; Strumberg, D.; Fasching, P.A.; Kreienberg, R.; et al. Lapatinib versus trastuzumab in combination with neoadjuvant anthracycline-taxane-based chemotherapy (GeparQuinto, GBG 44): A randomised phase 3 trial. Lancet Oncol. 2012, 13, 135–144. [Google Scholar] [CrossRef]
- Reinisch, M.; von Minckwitz, G.; Harbeck, N.; Janni, W.; Kümmel, S.; Kaufmann, M.; Elling, D.; Nekljudova, V.; Loibl, S. Side Effects of Standard Adjuvant and Neoadjuvant Chemotherapy Regimens According to Age Groups in Primary Breast Cancer. Breast Care 2013, 8, 60–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murthy, R.K.; Raghavendra, A.S.; Hess, K.R.; Fujii, T.; Lim, B.; Barcenas, C.H.; Zhang, H.; Chavez-Mac-Gregor, M.; Mittendorf, E.A.; Litton, J.K.; et al. Neoadjuvant Pertuzumab-containing Regimens Improve Pathologic Complete Response Rates in Stage II to III HER-2/neu-positive Breast Cancer: A Retrospective, Single Institution Experience. Clin. Breast Cancer 2018, 18, e1283–e1288. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.C.; Berry, D.; Carey, L.A.; Colleoni, M.; Dowsett, M.; Ellis, M.; Garber, J.E.; Mankoff, D.; Paik, S.; Pusztai, L.; et al. Research issues affecting preoperative systemic therapy for operable breast cancer. J. Clin. Oncol. 2008, 26, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Potti, A.; Dressman, H.K.; Bild, A.; Riedel, R.F.; Chan, G.; Sayer, R.; Cragun, J.; Cottrill, H.; Kelley, M.J.; Petersen, R.; et al. Genomic signatures to guide the use of chemotherapeutics. Nat. Med. 2006, 12, 1294–1300. [Google Scholar] [CrossRef]
- Zitvogel, L.; Apetoh, L.; Ghiringhelli, F.; Kroemer, G. Immunological aspects of cancer chemotherapy. Nat. Rev. 2008, 8, 59–73. [Google Scholar] [CrossRef]
- Chang, J.C.; Wooten, E.C.; Tsimelzon, A.; Hilsenbeck, S.G.; Gutierrez, M.C.; Elledge, R.; Mohsin, S.; Osborne, C.K.; Chamness, G.C.; Allred, D.C.; et al. Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet 2003, 362, 362–369. [Google Scholar] [CrossRef]
- Balko, J.M.; Cook, R.S.; Vaught, D.B.; Kuba, M.G.; Miller, T.W.; Bhola, N.E.; Sanders, M.E.; Granja-ingram, N.M.; Smith, J.J.; Ingrid, M. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat. Med. 2012, 18, 1052–1059. [Google Scholar] [CrossRef] [Green Version]
- Sohn, J.; Do, K.A.; Liu, S.; Chen, H.; Mills, G.B.; Hortobagyi, G.N.; Meric-Bernstam, F.; Gonzalez-Angulo, A.M. Functional proteomics characterization of residual triple-negative breast cancer after standard neoadjuvant chemotherapy. Ann. Oncol. 2013, 24, 2522–2526. [Google Scholar] [CrossRef]
- Satpathy, S.; Jaehnig, E.J.; Krug, K.; Kim, B.; Saltzman, A.B.; Chan, D.W.; Holloway, K.R.; Anurag, M.; Huang, C.; Singh, P.; et al. Microscaled proteogenomic methods for precision oncology. Nat. Commun. 2020, 11, 532. [Google Scholar] [CrossRef] [Green Version]
- Llombart-Cussac, A.; Cortés, J.; Paré, L.; Galván, P.; Bermejo, B.; Martínez, N.; Vidal, M.; Pernas, S.; López, R.; Muñoz, M.; et al. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): An open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 2017, 18, 545–554. [Google Scholar] [CrossRef]
- Lesurf, R.; Griffith, O.L.; Griffith, M.; Hundal, J.; Trani, L.; Watson, M.A.; Aft, R.; Ellis, M.J.; Ota, D.; Suman, V.J.; et al. Genomic characterization of HER2-positive breast cancer and response to neoadjuvant trastuzumab and chemotherapy-results from the ACOSOG Z1041 (Alliance) trial. Ann. Oncol. 2017, 28, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, D.; Venet, D.; Ignatiadis, M.; Azim, H.A., Jr.; Maetens, M.; Rothé, F.; Salgado, R.; Bradbury, I.; Pusztai, L.; Harbeck, N.; et al. RNA Sequencing to Predict Response to Neoadjuvant Anti-HER2 Therapy: A Secondary Analysis of the NeoALTTO Randomized Clinical Trial. JAMA Oncol. 2017, 3, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Bianchini, G.; Thomas, M.; Belousov, A.; Cheang, M.C.U.; Koehler, A.; Gómez, P.; Semiglazov, V.; Eiermann, W.; Tjulandin, S.; et al. Research-based PAM50 subtype predictor identifies higher responses and improved survival outcomes in HER2-positive breast cancer in the NOAH study. Clin. Cancer Res. 2014, 20, 511–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanioka, M.; Fan, C.; Parker, J.S.; Hoadley, K.A.; Hu, Z.; Li, Y.; Hyslop, T.M.; Pitcher, B.N.; Soloway, M.G.; Spears, P.A.; et al. Integrated Analysis of RNA and DNA from the Phase III Trial CALGB 40601 Identifies Predictors of Response to Trastuzumab-Based Neoadjuvant Chemotherapy in HER2-Positive Breast Cancer. Clin. Cancer Res. 2018, 24, 5292–5304. [Google Scholar] [CrossRef] [Green Version]
- Loi, S.; Michiels, S.; Salgado, R.; Sirtaine, N.; Jose, V.; Fumagalli, D.; Kellokumpu-Lehtinen, P.-L.; Bono, P.; Kataja, V.; Desmedt, C.; et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann. Oncol. 2014, 25, 1544–1550. [Google Scholar] [CrossRef]
- Nuciforo, P.; Pascual, T.; Cortés, J.; Llombart-Cussac, A.; Fasani, R.; Pare, L.; Oliveira, M.; Galvan, P.; Martínez, N.; Bermejo, B.; et al. A predictive model of pathologic response based on tumor cellularity and tumorinfiltrating lymphocytes (CelTIL) in HER2-positive breast cancer treated with chemo-free dual HER2 blockade. Ann. Oncol. 2018, 29, 170–177. [Google Scholar] [CrossRef]
- Varadan, V.; Gilmore, H.; Miskimen, K.L.; Tuck, D.; Parsai, S.; Awadallah, A.; Krop, I.E.; Winer, E.P.; Bossuyt, V.; Somlo, G.; et al. Immune Signatures Following Single Dose Trastuzumab Predict Pathologic Response to Preoperative Trastuzumab and Chemotherapy in HER2-Positive Early Breast Cancer. Clin. Cancer Res. 2016, 22, 3249–3259. [Google Scholar] [CrossRef] [Green Version]
- McNamara, K.L.; Caswell-Jin, J.L.; Joshi, R.; Ma, Z.; Kotler, E.; Bean, G.R.; Kriner, M.; Zhou, Z.; Hoang, M.; Beechem, J.; et al. Spatial proteomic characterization of HER2-positive breast tumors through neoadjuvant therapy predicts response. Nat. Cancer 2021, 2, 400–413. [Google Scholar] [CrossRef]
- Haugen, M.H.; Lingjærde, O.C.; Hedenfalk, I.; Garred, Ø.; Borgen, E.; Loman, N.; Hatschek, T.; Børresen-Dale, A.; Naume, B.; Mills, G.B.; et al. Protein Signature Predicts Response to Neoadjuvant Treatment With Chemotherapy and Bevacizumab in HER2-Negative Breast Cancers. JCO Precis. Oncol. 2021, 5, 286–306. [Google Scholar] [CrossRef]
- Chen, Y.; Li, H.; Liang, J.; Ger, L.; Chang, H.; Hsiao, M.; Calkins, M.J.; Cheng, H.; Chuang, J.; Lu, P. CTMP, a predictive biomarker for trastuzumab resistance in HER2-enriched breast cancer patient. Oncotarget 2017, 8, 29699–29710. [Google Scholar] [CrossRef] [Green Version]
- Lurie, Y.; Webb, M.; Cytter-Kuint, R.; Shteingart, S.; Lederkremer, G.Z. Non-invasive diagnosis of liver fibrosis and cirrhosis. World J. Gastroenterol. 2015, 21, 11567–11583. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Xu, P.; Gu, L.; Xu, Z.; Ge, W.; Li, Q.; Xu, F. Quantitative assessment of serum heat shock protein 27 for the diagnosis of epithelial ovarian cancer using targeted proteomics coupled with immunoaffinity enrichment. Clin. Chim. Acta 2019, 489, 96–102. [Google Scholar] [CrossRef]
- Yang, F.; Fu, Z.; Yang, M.; Sun, C.; Li, Y.; Chu, J.; Zhang, Y.; Li, W.; Huang, X.; Li, J.; et al. Expression pattern of microRNAs related with response to trastuzumab in breast cancer. J. Cell. Physiol. 2019, 234, 16102–16113. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, S.; Giordano, A.; Gao, H.; Cohen, E.N.; Tin, S.; Wu, Q.; Garza, R.J.; Debeb, B.G.; Alvarez, R.H.; Valero, V.; et al. High serum miR-19a levels are associated with inflammatory breast cancer and are predictive of favorable clinical outcome in patients with metastatic HER2+ inflammatory breast cancer. PLoS ONE 2014, 9, e83113. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, J.; Chen, J.; Wang, H.; Yang, L.; Chen, F.; Fan, S.; Wang, J.; Shao, B.; Yin, D.; et al. A serum microRNA signature predicts trastuzumab benefit in HER2-positive metastatic breast cancer patients. Nat. Commun. 2018, 9, 1614. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yang, F.; Zhang, Y.; Chu, J.; Wang, J.; Wang, Y.; Zhang, Y.; Li, J.; Li, Y.; Fan, R.; et al. tRNA-Derived Fragments as Novel Predictive Biomarkers for Trastuzumab-Resistant Breast Cancer. Cell. Physiol. Biochem. 2018, 49, 419–431. [Google Scholar] [CrossRef] [PubMed]
- Fornier, M.N.; Seidman, A.D.; Schwartz, M.K.; Ghani, F.; Thiel, R.; Norton, L.; Hudis, C. Serum HER2 extracellular domain in metastatic breast cancer patients treated with weekly trastuzumab and paclitaxel: Association with HER2 status by immunohistochemistry and fluorescence in situ hybridization and with response rate. Ann. Oncol. 2005, 16, 234–239. [Google Scholar] [CrossRef]
- Ho, D.; Huang, J.; Chapman, J.W.; Leitzel, K.; Ali, S.M.; Shepherd, L.; Parulekar, W.R.; Ellis, C.E.; Crescnzo, R.J.; Zhu, L.; et al. Impact of serum HER2, TIMP-1, and CAIX on outcome for HER2+ metastatic breast cancer patients: CCTG MA.31 (lapatinib vs. trastuzumab). Breast Cancer Res. Treat. 2017, 164, 571–580. [Google Scholar] [CrossRef]
- Nami, B.; Wang, Z. HER2 in breast Cancer stemness: A negative feedback loop towards trastuzumab resistance. Cancers 2017, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Lu, Q.; Liang, J.; Xia, Y.; Zhang, W.; Hu, B.; Shang, F.; Ji, Y.; Wang, J.; Wang, Q.; et al. High Plasma Fibrinogen is Correlated With Poor Response to Trastuzumab Treatment in HER2 Positive Breast Cancer. Medicine 2015, 94, e481. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, X.; Wu, D.; Gao, H.; Jiang, J.; Yang, Y.; Wu, J.; Gao, Q.; Wang, J.; Jiang, Z.; et al. Circulating proteomic panels for diagnosis and risk stratification of acute-on-chronic liver failure in patients with viral hepatitis B. Theranostics 2019, 9, 1200–1214. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Yang, Y.; Wang, J.; Zhang, L.; Wang, F.; He, A. Variability of serum novel serum peptide biomarkers correlates with the disease states of multiple mieloma. Clin. Proteom. 2019, 16, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tremlett, H.; Dai, D.L.; Hollander, Z.; Kapanen, A.; Aziz, T.; Wilson-McManus, J.E.; Tebbutt, S.J.; Borchers, C.H.; Oger, J.; Cohen Freue, G.V. Serum proteomics in multiple sclerosis disease progression. J. Proteom. 2015, 118, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Mysona, D.; Pyrzak, A.; Purohit, S.; Zhi, W.; Sharma, A.; Tran, L.; Tran, P.; Bai, S.; Rungruang, B.; Ghamande, S.; et al. A combined score of clinical factors and serum proteins can predict time to recurrence in high grade serous ovarian cancer. Gynecol. Oncol. 2019, 152, 574–580. [Google Scholar] [CrossRef]
- Yang, T.; Fu, Z.; Zhang, Y.; Wang, M.; Mao, C.; Ge, W. Serum proteomics analysis of candidate predictive biomarker panel for the diagnosis of trastuzumab-based therapy resistant breast cancer. Biomed. Pharmacother. 2020, 129, 110465. [Google Scholar] [CrossRef]
- Jia, L.; Lu, Y.; Shao, J.; Liang, X.J.; Xu, Y. Nanoproteomics: A new sprout from emerging links between nanotechnology and proteomics. Trends Biotechnol. 2013, 31, 99–107. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.P.; Castro López, A.; Bravo, S.B.; Vázquez-Estévez, S.; Acea-Nebril, B.; Núñez, C. Proteomic analysis of the bio-corona formed on the surface of (Au, Ag, Pt)-nanoparticles in human serum. Colloids Surf. B Biointerfaces 2019, 177, 141–148. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.P.; Castro López, A.; García Vence, M.; Vázquez-Estévez, S.; Acea-Nebril, B.; Calatayud, D.G.; Jardiel, T.; Bravo, S.B.; Núñez, C. Proteomic investigation on bio-corona of Au, Ag and Fe nanoparticles for the discovery of triple negative breast cancer serum protein biomarkers. J. Proteom. 2020, 212, 103581. [Google Scholar] [CrossRef] [PubMed]
- Chantada-Vázquez, M.P.; García-Vence, M.; Vázquez-Estévez, S.; Bravo, S.B.; Núñez, C. Identification of a Profile of Neutrophil-Derived Granule Proteins in the Surface of Gold Nanoparticles after Their Interaction with Human Breast Cancer Sera. Nanomaterials 2020, 10, 1223. [Google Scholar] [CrossRef]
- Chantada-Vázquez, M.D.P.; López, A.C.; García-Vence, M.; Acea-Nebril, B.; Bravo, S.B.; Núñez, C. Protein corona gold nanoparticles fingerprinting reveals a profile of blood coagulation proteins in the serum of Her2-overexpressing breast cancer patients. Int. J. Mol. Sci. 2020, 21, 8449. [Google Scholar] [CrossRef]
- García-Vence, M.; Chantada-Vázquez, M.D.P.; Cameselle-Teijeiro, J.M.; Bravo, S.B.; Núñez, C. A Novel nanoproteomic approach for the identification of molecular targets associated with thyroid tumors. Nanomaterials 2020, 10, 2370. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Gómez, B.; López-Cortés, R.; Casas-Nebra, F.J.; Vázquez-Estévez, S.; Pérez-Fentes, D.; Chantada-Vázquez, M.D.P.; Bravo, S.B.; Núñez, C. Detection of Circulating Serum Protein Biomarkers of Non-Muscle Invasive Bladder Cancer after Protein Corona-Silver Nanoparticles Analysis by SWATH-MS. Nanomaterials 2021, 11, 2384. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.W.; Yan, Y.; Caruso, F.; Nice, E.C. Emerging techniques in proteomics for probing nano-bio interactions. ACS Nano 2012, 6, 10438–10448. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Wang, Y.; Song, Z.; Feng, Y.; Chen, Y.; Zhang, D.; Feng, L. The Basic Properties of Gold Nanoparticles and their Applications in Tumor Diagnosis and Treatment. Int. J. Mol. Sci. 2020, 21, 2480. [Google Scholar] [CrossRef] [Green Version]
- Jeyaraj, M.; Gurunathan, S.; Qasim, M.; Kang, M.-H.; Kim, J.-H. A Comprehensive Review on the Synthesis, Characterization, and Biomedical Application of Platinum Nanoparticles. Nanomaterials 2019, 9, 1719. [Google Scholar] [CrossRef] [Green Version]
- Edge, S.B.; Byrd, D.R.; Compton, C.C.; Fritz, A.G.; Greene, F.L.; Trotti, A. American Joint Committee on Cancer (AJCC) Cancer Staging Manual, 7th ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Warder, S.E.; Tucker, L.A.; Strelitzer, T.J.; McKeegan, E.M.; Meuth, J.L.; Jung, P.M.; Saraf, A.; Singh, B.; Lai-Zhang, J.; Gagne, G.; et al. Reducing agent-mediated precipitation of high-abundance plasma proteins. Anal. Biochem. 2009, 387, 184–193. [Google Scholar] [CrossRef]
- Fernández, C.; Santos, H.M.; Ruíz-Romero, C.; Blanco, F.J.; Capelo-Martínez, J.L. A comparison of depletion versus equalization for reducing high-abundance proteins in human serum. Electrophoresis 2011, 32, 2966–2974. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Jensen, O.N.; Podtelejnikov, A.V.; Neubauer, G.; Mortensen, P.; Mann, M. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 1996, 24, 893–896. [Google Scholar] [CrossRef] [Green Version]
- Shilov, I.V.; Seymour, S.L.; Patel, A.A.; Loboda, A.; Tang, W.H.; Keating, S.P.; Hunter, C.L.; Nuwaysir, L.M.; Schaeffer, D.A. The paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell Proteom. 2007, 6, 1638–1655. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.H.; Shilov, I.V.; Seymour, S.L. Nonlinear fitting method for determining local false discovery rates from decoy database searches. J. Proteome Res. 2008, 7, 3661–3667. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. pROC: An open-source package for R and Sþ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, T. An introduction to ROC analysis. Pattern Recogn. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Tirumalai, R.S.; Chan, K.C.; Prieto, D.A.; Issaq, H.J.; Conrads, T.P.; Veenstra, T.D. Characterization of the low molecular weight human serum proteome. Mol. Cell. Proteom. 2003, 2, 1096–1103. [Google Scholar] [CrossRef] [Green Version]
- Cui, R.; Zhang, P.; Li, Y. Role of Carboxypeptidase N Invasion and Migration in Breast Cancer. Anti-Cancer Agents Med. Chem. 2016, 16, 1198–1202. [Google Scholar] [CrossRef]
- Xu, X.; Wan, J.; Yuan, L.; Ba, J.; Feng, P.; Long, W.; Huang, H.; Liu, P.; Cai, Y.; Liu, M.; et al. Serum levels of apolipoprotein E correlates with disease progression and poor prognosis in breast cancer. Tumor Biol. 2016, 37, 15959–15966. [Google Scholar] [CrossRef]
- Zhou, Y.; Luo, G. Apolipoproteins, as the carrier proteins for lipids, are involved in the development of breast cancer. Clin. Transl. Oncol. 2020, 22, 1952–1962. [Google Scholar] [CrossRef]
- Okuturlar, Y.; Gunaldi, M.; Kocoglu, H.; Hursitoglu, M.; Gedikbasi, A.; Acarer, D.; Harmankaya, O.; Kumbasar, A. Serum paraoxonase and arylesterase can be useful markers to predict neoadjuvant chemotherapy requirement in patients with breast cancer. J. Cancer Res. Ther. 2018, 14, S362–S367. [Google Scholar]
- Lima, L.G.; Monteiro, R.Q. Activation of blood coagulation in cancer: Implications for tumour progression. Biosci. Rep. 2013, 33, e00064. [Google Scholar] [CrossRef]
- Metelli, A.; Wu, B.X.; Riesenberg, B.; Guglietta, S.; Huck, J.D.; Mills, C.; Li, A.; Rachidi, S.; Krieg, C.; Rubinstein, M.P.; et al. Thrombin contributes to cancer immune evasion via proteolysis of platelet-bound GARP to activate LTGF-β. Sci. Transl. Med. 2020, 12, eaay4860. [Google Scholar] [CrossRef] [PubMed]
- Hisada, Y.; Mackman, N. Cancer-associated pathways and biomarkers of venous thrombosis. Blood 2017, 130, 1499–1506. [Google Scholar] [CrossRef] [PubMed]
- Ruf, W.; Rothmeier, A.S.; Graf, C. Targeting clotting proteins in cancer therapy—progress and challenges. Thromb. Res. 2016, 140, S1–S7. [Google Scholar] [CrossRef] [Green Version]
- Zelvyte, I.; Sjögren, H.O.; Janciauskiene, S. Effects of native and cleaved forms of alpha1-antitrypsin on ME 1477 tumor cell functional activity. Cancer Detect. Prev. 2002, 26, 256–265. [Google Scholar] [CrossRef]
- Lal, I.; Dittus, K.; Holmes, C.E. Platelets, coagulation and fibrinolysis in breast cancer progression. Breast Cancer Res. 2013, 15, 207. [Google Scholar] [CrossRef] [Green Version]
- Tas, F.; Kilic, L.; Duranyildiz, D. Coagulation tests show significant differences in patients with breast cancer. Tumor Biol. 2014, 35, 5985–5992. [Google Scholar] [CrossRef]
- Yigit, E.; Gönüllü, G.; Yücel, I.; Turgut, M.; Erdem, D.; Cakar, B. Relation between hemostatic parameters and prognostic/predictive factors in breast cancer. Eur. J. Intern. Med. 2008, 19, 602–607. [Google Scholar] [CrossRef]
- Zacharski, L.R.; Henderson, W.G.; Rickles, F.R.; Forman, W.B.; Cornell, C.J., Jr.; Forcier, A.J.; Edwards, R.L.; Headley, E.; Kim, S.-H.; O’Donnell, J.F.; et al. Effect of warfarin anticoagulation on survival in carcinoma of the lung, colon, head and neck, and prostate. Cancer 1984, 53, 2046–2052. [Google Scholar] [CrossRef]
- Kolev, M.; Maciej, M. Targeting complement-mediated immunoregulation for cancer immunotherapy. Semin. Immunol. 2018, 37, 85–97. [Google Scholar] [CrossRef]
- Habermann, J.K.; Roblick, U.J.; Luke, B.T.; Prieto, D.A.; Finlay, W.J.; Podust, V.N.; Roman, J.M.; Oevermann, E.; Schiedeck, T.; Homann, N.; et al. Increased serum levels of complement C3a anaphylatoxin indicate the presence of colorectal tumors. Gastroenterology 2006, 131, 1020–1029. [Google Scholar] [CrossRef] [Green Version]
- Ward, D.G.; Suggett, N.; Cheng, Y.; Wei, W.; Johnson, H.; Billingham, L.J.; Ismail, T.; Wakelam, M.J.; Johnson, P.J.; Martin, A. Identification of serum biomarkers for colon cancer by proteomic analysis. Br. J. Cancer 2006, 94, 1898–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakshatri, H.; Qi, Q.; You, J.; Kerry, B.; Schneider, B.; Zon, R.; Buck, C.; Regnier, F.; Wang, M. Intrinsic subtype-associated changes in the plasma proteome in breast cancer. Proteom. Clin. Appl. 2009, 3, 1305–1313. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, A.; Esterni, B.; Bertucci, F.; Sauvan, R.; Chabannon, C.; Cubizolles, M.; Bardou, V.J.; Houvenaegel, G.; Jacquemier, J.; Granjeaud, S.; et al. Postoperative serum proteomic profiles may predict metastatic relapse in high-risk primary breast cancer patients receiving adjuvant chemotherapy. Oncogene 2006, 25, 981–998. [Google Scholar] [CrossRef]
- Michlmayr, A.; Bachleitner-Hofmann, T.; Baumann, S.; Marchetti-Deschmann, M.; Rech-Weichselbraun, I.; Burghuber, C.; Pluschnig, U.; Bartsch, R.; Graf, A.; Greil, R.; et al. Modulation of plasma complement by the initial dose of epirubicin/docetaxel therapy in breast cancer and its predictive value. Br. J. Cancer 2010, 103, 1201–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricklin, D.; Mastellos, D.C.; Reis, E.S.; Lambris, J.D. The renaissance of complement therapeutics. Nat. Rev. Nephrol. 2018, 14, 26–47. [Google Scholar] [CrossRef] [Green Version]
- Surace, L.; Lysenko, V.; Fontana, A.O.; Cecconi, V.; Janssen, H.; Bicvic, A.; Okoniewski, M.; Pruschy, M.; Dummer, R.; Neefjes, J.; et al. Complement is a central mediator of radiotherapy-induced tumor-specific immunity and clinical response. Immunity 2015, 42, 767–777. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Zhao, Q.; Liao, J.-Y.; Song, E.; Xia, Q.; Pan, J.; Li, Y.; Li, J.; Zhou, B.; Ye, Y.; et al. Complement Signals Determine Opposite Effects of B Cells in Chemotherapy-Induced Immunity. Cell 2020, 180, 1081–1097. [Google Scholar] [CrossRef]
- Cibeira, G.H.; Giacomazzi, J.; Aguiar, E.; Schneider, S.; Ettrich, B.; De Souza, C.I.; Camey, S.; Caleffi, M.; Weber, B.; Ashton-Prolla, P.; et al. Apolipoprotein E genetic polymorphism, serum lipoprotein levels and breast cancer risk: A case-control study. Mol. Clin. Oncol. 2014, 2, 1009–1015. [Google Scholar] [CrossRef]
- Liu, J.-X.; Yuan, Q.; Min, Y.-L.; He, Y.; Xu, Q.-H.; Li, B.; Shi, W.-Q.; Lin, Q.; Li, Q.-H.; Zhu, P.-W.; et al. Apolipoprotein A1 and B as risk factors for development of intraocular metastasis in patients with breast cancer. Cancer Manag. Res. 2019, 11, 2881–2888. [Google Scholar] [CrossRef] [Green Version]
- Oddoze, C.; Michel, B.F. Apolipoprotein Ea4 allele predicts a better response to donepezil therapy in Alzheimer’s disease. Alzheimer’s Rep. 2000, 3, 213–216. [Google Scholar]
- Del Boccio, P.; Perrotti, F.; Rossi, C.; Cicalini, I.; Di Santo, S.; Zucchelli, M.; Sacchetta, P.; Genovesi, D.; Pieragostino, D. Serum lipidomic study reveals potential early biomarkers for predicting response to chemoradiation therapy in advanced rectal cancer: A pilot study. Adv. Radiat. Oncol. 2017, 2, 118–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Chen, C.; Zeng, Z.; Wang, Q.; Jiang, W.; Gao, Y.; Chang, H. Serum Apolipoprotein A-I Predicts Response of Rectal Cancer to Neoadjuvant Chemoradiotherapy. Cancer Manag. Res. 2021, 13, 2623–2631. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Shi, L.; Liu, Q.; Dong, R.; Zhang, Q.; Yang, S.; Fan, Y.; Yang, H.; Wu, P.; Yu, J.; et al. Discovery and identification of potential biomarkers of papillary thyroid carcinoma. Mol. Cancer 2009, 8, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, K.; Okusaka, T.; Felix, K.; Nakamori, S.; Sata, N.; Nagai, H.; Ioka, T.; Tsuchida, A.; Shimahara, T.; Shimahara, M.; et al. Altered plasma apolipoprotein modifications in patients with pancreatic cancer: Protein characterization and multi-institutional validation. PLoS ONE 2012, 7, e46908. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Yang, H.; Duan, X.; Li, L.; Sun, L.; Li, Q.; Zhang, J. Apolipoproteins as Differentiating and Predictive Markers for Assessing Clinical Outcomes in Patients with Small Cell Lung Cancer. Yonsei Med. J. 2016, 57, 549–556. [Google Scholar] [CrossRef]
- Kardassis, D.; Pardali, K.; Zannis, V.I. SMAD proteins transactivate the human ApoCIII promoter by interacting physically and functionally with hepatocyte nuclear factor 4. J. Biol. Chem. 2000, 275, 41405–41414. [Google Scholar] [CrossRef] [Green Version]
- Fraser, J.D.; Martinez, V.; Straney, R.; Briggs, M.R. DNA binding and transcription activation specificity of hepatocyte nuclear factor 4. Nucleic Acids Res. 1998, 26, 2702–2707. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, L.M.; Diez-Itza, I.; Vizoso, F.; Lopez-Otin, C. Cholesterol and apolipoprotein D in gross cystic disease of the breast. Clin Chem. 1992, 38, 695–698. [Google Scholar] [CrossRef]
- Diez-Itza, I.; Vizoso, F.; Merino, A.M.; Sanchez, L.M.; Tolivia, J.; Fernandez, J.; Ruibal, A.; López-Otín, C. Expression and prognostic signifcance of apolipoprotein D in breast cancer. Am. J. Pathol. 1994, 144, 310–320. [Google Scholar]
- Sivaraman, V.S.; Wang, H.; Nuovo, G.J.; Malbon, C.C. Hyperexpression of mitogen-activated protein kinase in human breast cancer. J. Clin. Investig. 1997, 99, 1478–1483. [Google Scholar] [CrossRef]
- Sarjeant, J.M.; Lawrie, A.; Kinnear, C.; Yablonsky, S.; Leung, W.; Massaeli, H.; Prichett, W.; Veinot, J.P.; Rassart, E.; Rabinovitch, M. Apolipoprotein D inhibits platelet-derived growth factor-BB-induced vascular smooth muscle cell proliferated by preventing translocation of phosphorylated extracellular signal regulated kinase 1/2 to the nucleus. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 2172–2177. [Google Scholar] [CrossRef]
- Romano, M.; Claria, J. Cyclooxygenase-2 and 5-lipoxygenase converging functions on cell proliferation and tumor angiogenesis: Implications for cancer therapy. FASEB J. 2003, 17, 1986–1995. [Google Scholar] [CrossRef] [Green Version]
- Matsuyama, M.; Yoshimura, R.; Mitsuhashi, M.; Tsuchida, K.; Takemoto, Y.; Kawahito, Y.; Sano, H.; Nakatani, T. 5-Lipoxygenase inhibitors attenuate growth of human renal cell carcinoma and induce apoptosis through arachidonic acid pathway. Oncol. Rep. 2005, 14, 73–79. [Google Scholar] [PubMed]
- Soiland, H.; Soreide, K.; Janssen, E.A.; Korner, H.; Baak, J.P.; Soreide, J.A. Emerging concepts of apolipoprotein D with possible implications for breast cancer. Cell. Oncol. 2007, 29, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Lamelas, M.L.; Vazquez, J.; Enguita, M.I.; Rodriguez, J.C.; Gonzalez, L.O.; Merino, A.M.; Vizoso, F. Apolipoprotein D expression in metastasic lymph nodes of breast cancer. Int. J. Surg. Investig. 2000, 2, 285–293. [Google Scholar] [PubMed]
- Carreno, G.; Del Casar, J.M.; Corte, M.D.; Gonzalez, L.O.; Bongera, M.; Merino, A.M.; Juan, G.; Obregón, R.; Martínez, E.; Vizoso, F.J. Local recurrence after mastectomy for breast cancer: Analysis of clinicopathological, biological and prognostic characteristics. Breast Cancer Res. Treat. 2007, 102, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Do Carmo, S.; Levros, L.C., Jr.; Rassart, E. Modulation of apolipoprotein D expression and translocation under specific stress conditions. Biochim. Biophys. Acta 2007, 1773, 954–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harding, C.; Osundeko, O.; Tetlow, L.; Faragher, E.B.; Howell, A.; Bundred, N.J. Hormonally-regulated proteins in breast secretions are markers of target organ sensitivity. Br. J. Cancer. 2000, 82, 354–360. [Google Scholar] [CrossRef]
- Simard, J.; Dauvois, S.; Haagensen, D.E.; Levesque, C.; Merand, Y.; Labrie, F. Regulation of progesterone-binding breast cyst protein GCDFP-24 secretion by estrogens and androgens in human breast cancer cells: A new marker of steroid action in breast cancer. Endocrinology 1990, 126, 3223–3231. [Google Scholar] [CrossRef]
- Beelen, K.; Zwart, W.; Linn, S.C. Can predictive biomarkers in breast cancer guide adjuvant endocrine therapy? Nat. Rev. Clin. Oncol. 2012, 9, 529–541. [Google Scholar] [CrossRef]
- Soreide, J.A.; Lea, O.A.; Anda, O.; Skarstein, A.; Varhaug, J.E.; Kvinnsland, S. Progesterone-binding cyst protein (PBCP) in operable breast cancer: Correlations with prognostic factors and predictive value for effect of adjuvant tamoxifen treatment. Anticancer Res. 1991, 11, 601–605. [Google Scholar] [PubMed]
- Kiselar, J.G.; Janmey, P.A.; Almo, S.C.; Chance, M.R. Visualizing the Ca2+-dependent activation of gelsolin by using synchrotron footprinting. Proc. Natl. Acad. Sci. USA 2003, 100, 3942–3947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.L.; Stossel, T.P. Control of cytoplasmic actin gel–sol transformation by gelsolin, a calcium-dependent regulatory protein. Nature 1979, 281, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Li, G.H.; Arora, P.D.; Chen, Y.; McCulloch, C.A.; Liu, P. Multifunctional roles of gelsolin in health and diseases. Med. Res. Rev. 2012, 32, 999–1025. [Google Scholar] [CrossRef] [PubMed]
- Baig, R.M.; Mahjabeen, I.; Sabir, M.; Masood, N.; Ali, K.; Malik, F.A.; Kayani, M.A. Mutational spectrum of gelsolin and its down regulation is associated with breast cancer. Dis. Markers 2013, 34, 71–80. [Google Scholar] [CrossRef]
- Visapää, H.; Bui, M.; Huang, Y.; Seligson, D.; Tsai, H.; Pantuck, A.; Figlin, R.; Rao, J.Y.; Belldegrun, A.; Horvath, S.; et al. Correlation of Ki-67 and gelsolin expression to clinical outcome in renal clear cell carcinoma. Urology 2003, 61, 845–850. [Google Scholar] [CrossRef]
- Litwin, M.; Nowak, D.; Mazur, A.J.; Baczyńska, D.; Mannherz, H.G.; Malicka-Błaszkiewicz, M. Gelsolin affects the migratory ability of human colon adenocarcinoma and melanoma cells. Life Sci. 2012, 90, 851–861. [Google Scholar] [CrossRef]
- Radwanska, A.; Litwin, M.; Nowak, D.; Baczynska, D.; Wegrowski, Y.; Maquart, F.X.; Malicka-Blaszkiewicz, M. Overexpression of lumican affects the migration of human colon cancer cells through up-regulation of gelsolin and filamentous actin reorganization. Exp. Cell Res. 2012, 318, 2312–2323. [Google Scholar] [CrossRef]
- Deng, R.; Hao, J.; Han, W.; Ni, Y.; Huang, X.; Hu, Q. Gelsolin regulates proliferation, apoptosis, migration and invasion in human oral carcinoma cells. Oncol. Lett. 2015, 9, 2129–2134. [Google Scholar] [CrossRef]
- Shieh, D.B.; Chen, I.W.; Wei, T.Y.; Shao, C.Y.; Chang, H.J.; Chung, C.H.; Wong, T.Y.; Jin, Y.T. Tissue expression of gelsolin in oral carcinogenesis progression and its clinicopathological implications. Oral Oncol. 2006, 42, 599–606. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.G.; Huang, R.; Yao, C.; Li, S.; Yang, W.; Yang, N.; Huang, R. Concurrent downregulation of Egr-1 and gelsolin in the majority of human breast cancer cells. Cancer Genom. Proteom. 2007, 4, 377–385. [Google Scholar]
- Thor, A.D.; Edgerton, S.M.; Liu, S.; Moore, D.H., 2nd; Kwiatkowski, D.J. Gelsolin as a negative prognostic factor and effector of motility in erbB-2-positive epidermal growth factor receptor-positive breast cancers. Clin. Cancer Res. 2001, 7, 2415–2424. [Google Scholar] [PubMed]
- Winston, J.S.; Asch, H.L.; Zhang, P.J.; Edge, S.B.; Hyland, A.; Asch, B.B. Downregulation of gelsolin correlates with the progression to breast carcinoma. Breast Cancer Res. Treat. 2001, 65, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Asch, H.L.; Winston, J.S.; Edge, S.B.; Stomper, P.C.; Asch, B.B. Down-regulation of gelsolin expression in human breast ductal carcinoma in situ with and without invasion. Breast Cancer Res. Treat. 1999, 55, 179–188. [Google Scholar] [CrossRef]
- Asch, H.L.; Head, K.; Dong, Y.; Natoli, F.; Winston, J.S.; Connolly, J.L.; Asch, B.B. Widespread loss of gelsolin in breast cancers of humans, mice, and rats. Cancer Res. 1996, 56, 4841–4845. [Google Scholar] [PubMed]
- Wang, P.W.; Abedini, M.R.; Yang, L.X.; Ding, A.A.; Figeys, D.; Chang, J.Y.; Tsang, B.K.; Shieh, D.-B. Gelsolin regulates cisplatin sensitivity in human head-and-neck cancer. Int. J. Cancer. 2014, 135, 2760–2769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abedini, M.R.; Wang, P.W.; Huang, Y.F.; Cao, M.; Chou, C.Y.; Shieh, D.B.; Tsang, B.K. Cell fate regulation by gelsolin in human gynecologic cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 14442–14447. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.; Hellwig, B.; Hammad, S.; Othman, A.; Lohr, M.; Chen, Z.; Boehm, D.; Gebhard, S.; Petry, I.; Lebrecht, A.; et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin κ C as a compatible prognostic marker in human solid tumors. Clin. Cancer Res. 2012, 18, 2695–2703. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Gerhold-Ay, A.; Gebhard, S.; Boehm, D.; Solbach, C.; Lebrecht, A.; Battista, M.; Sicking, I.; Cotarelo, C.; Cadenas, C.; et al. Immunoglobulin kappa C predicts overall survival in node-negative breast cancer. PLoS ONE 2012, 7, e44741. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.; Böhm, D.; von Törne, C.; Steiner, E.; Puhl, A.; Pilch, H.; Lehr, H.-A.; Hengstler, J.; Kölbl, H.; Gehrmann, M. The humoral immune system has a key prognostic impact in nodenegative breast cancer. Cancer Res. 2008, 68, 5405–5413. [Google Scholar] [CrossRef] [Green Version]
- Heimes, A.; Krämer, H.; Härtner, F.; Almstedt, K.; Krajnak, S.; Battista, M.J.; Brenner, W.; Hasenburg, A.; Schmidt, M. Prognostic Impact of Immunoglobulin Kappa C in Breast Cancer Patients Treated with Adjuvant Chemotherapy. Breast Care 2021, 16, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L.; Ferrone, S. For Breast Cancer Prognosis, Immunoglobulin Kappa Chain Surfaces to the Top. Clin. Cancer Res. 2012, 18, 2417–2419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maehara, N.; Arai, S.; Mori, M.; Iwamura, Y.; Kurokawa, J.; Kai, T.; Kusunoki, S.; Taniguchi, K.; Ikeda, K.; Ohara, O.; et al. Circulating AIM prevents hepatocellular carcinoma through complement activation. Cell Rep. 2014, 9, 61–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.-K.; Tsai, C.-H.; Liu, Y.-W.; Lai, C.-C.; Huang, C.-C.; Sheen-Chen, S.-M. Afamin expression in breast cancer. Asian J. Surg. 2020, 43, 750–754. [Google Scholar] [CrossRef]
- Melmer, A.; Fineder, L.; Lamina, C.; Kollerits, B.; Dieplinger, B.; Braicu, I.; Sehouli, J.; Cadron, I.; Vergote, I.; Mahner, S.; et al. Plasma concentrations of the vitamin E-binding protein afamin are associated with overall and progression-free survival and platinum sensitivity in serous ovarian cancer—A study by the OVCAD consortium. Gynecol. Oncol. 2013, 128, 38–43. [Google Scholar] [CrossRef]
- Li, Q.; Lin, Y.; Li, Q.; Lin, Y. Evaluation of Ficolin-3 as a Potential Prognostic Serum Biomarker in Chinese Patients with Esophageal Cancer. Genet. Test. Mol. Biomark. 2019, 23, 565–572. [Google Scholar] [CrossRef]
- Sahar, T.; Nigam, A.; Anjum, S.; Gupta, N.; Wajid, S. Secretome Profiling and Computational Biology of Human Leiomyoma Samples Unravel Molecular Signatures with Potential for Diagnostic and Therapeutic Interventions. Reprod. Sci. 2021, 28, 2672–2684. [Google Scholar] [CrossRef]
- Muranjan, M.; Nussenzweig, V.; Tomlinson, S. Characterization of the human serum trypanosome toxin, haptoglobin-related protein. J. Biol. Chem. 1998, 273, 3884–3887. [Google Scholar] [CrossRef] [Green Version]






| Pat. No. | Age | Type | Tumor Size | T-Stage | N-Stage | ER | PR | HER-2 | Grading | Response Group |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 61 | Ductal | 3.4 | 2 | − | + | + | A | 1 | NR |
| 2 | 39 | Ductal | 2.6 | 2 | + | + | + | A | 1 | NR |
| 3 | 55 | Ductal | 2.5 | 2 | + | − | − | A | 2 | NR |
| 4 | 58 | Ductal | 2.4 | 2 | − | − | − | A | 2 | NR |
| 5 | 43 | Ductal | 2.4 | 2 | − | + | − | A | 2 | R |
| 6 | 36 | Ductal | 3.5 | 2 | − | + | + | A | 2 | R |
| 7 | 62 | Ductal | 3.2 | 2 | − | + | + | A | 3 | R |
| 8 | 64 | Ductal | 3 | 2 | + | + | − | A | 2 | R |
| 9 | 70 | Ductal | 2.4 | 2 | − | − | − | A | 3 | R |
| 10 | 44 | Ductal | 5.5 | 3 | − | − | − | A | 2 | R |
| Fraction | Number of Proteins Identified | ||||
|---|---|---|---|---|---|
| Total | Common | ||||
| Classification | Without NPs (method 1) | With AuNPs (method 2) | With PtNPs (method 3) | ||
| Responders (n = 6) | 129 | 61 | 56 | 43 | |
| Non-responders (n = 4) | 138 | 100 | 61 | 54 | |
 | |||||
| Protein Name | UniProt Name | Entry Name | Gene | Responders | Non-Responders |
|---|---|---|---|---|---|
| Apolipoprotein C-III | P02656 | APOC3_HUMAN | APOC3 | X | |
| Gelsolin | P06396 | GELS_HUMAN | GSN | X | |
| Immunoglobulin kappa constant | P01834 | IGKC_HUMAN | IGKC | X | |
| Immunoglobulin lambda-like polypeptide 5 | B9A064 | IGLL5_HUMAN | IGLL5 | X | |
| CD5 antigen-like | O43866 | CD5L_HUMAN | CD5L | X | |
| Afamin | P43652 | AFAM_HUMAN | AFM | X | |
| Plasminogen | P00747 | PLMN_HUMAN | PLG | X | |
| Ficolin-3 | O75636 | FCN3_HUMAN | FCN3 | X | |
| Complement factor H | P08603 | CFAH_HUMAN | CFH | X | |
| Complement factor H-related protein 1 | Q03591 | FHR1_HUMAN | CFHR1 | X | |
| Alpha-1-antitrypsin | P01009 | A1AT_HUMAN | SERPINA1 | X | |
| C4b-binding protein alpha chain | P04003 | C4BPA_HUMAN | C4BPA | X | |
| Complement factor I | P05156 | CFAI_HUMAN | CFI | X | |
| Complement C5 | P01031 | CO5_HUMAN | C5 | X | |
| Apolipoprotein D | P05090 | APOD_HUMAN | APOD | X | |
| Haptoglobin-related protein | P00739 | HPTR_HUMAN | HPR | X | |
| Prothrombin | P00734 | THRB_HUMAN | F2 | X | |
| Serum paraoxonase/arylesterase 1 | P27169 | PON1_HUMAN | PON1 | X | X |
| Immunoglobulin heavy constant gamma 1 | P01857 | IGHG1_HUMAN | IGHG1 | X | X |
| Inter-alpha-trypsin inhibitor heavy chain H3 | Q06033 | ITIH3_HUMAN | ITIH3 | X | X |
| Kininogen-1 | P01042 | KNG1_HUMAN | KNG1 | X | X |
| Plasma protease C1 inhibitor | P05155 | IC1_HUMAN | SERPING1 | X | X |
| Inter-alpha-trypsin inhibitor heavy chain H2 | P19823 | ITIH2_HUMAN | ITIH2 | X | X |
| Vitronectin | P04004 | VTNC_HUMAN | VTN | X | X |
| Vitamin D-binding protein | P02774 | VTDB_HUMAN | GC | X | X |
| Inter-alpha-trypsin inhibitor heavy chain H1 | P19827 | ITIH1_HUMAN | ITIH1 | X | X |
| Complement C1q subcomponent subunit C | P02747 | C1QC_HUMAN | C1QC | X | X |
| Antithrombin-III | P01008 | ANT3_HUMAN | SERPINC1 | X | X |
| Fibronectin | P02751 | FINC_HUMAN | FN1 | X | X |
| Apolipoprotein A-I | P02647 | APOA1_HUMAN | APOA1 | X | X |
| Complement C2 | P06681 | CO2_HUMAN | C2 | X | X |
| Hemopexin | P02790 | HEMO_HUMAN | HPX | X | X |
| Apolipoprotein E | P02649 | APOE_HUMAN | APOE | X | X |
| Immunoglobulin heavy constant alpha 1 | P01876 | IGHA1_HUMAN | IGHA1 | X | X |
| N-acetylmuramoyl-L-alanine amidase | Q96PD5 | PGRP2_HUMAN | PGLYRP2 | X | X |
| Haptoglobin | P00738 | HPT_HUMAN | HPT | X | X |
| Alpha-2-macroglobulin | P01023 | A2MG_HUMAN | A2M | X | X |
| Vitamin K-dependent protein S | P07225 | PROS_HUMAN | PROS1 | X | X |
| Immunoglobulin heavy constant mu | P01871 | IGHM_HUMAN | IGHM | X | X |
| Serotransferrin | P02787 | TRFE_HUMAN | TF | X | X |
| Clusterin | P10909 | CLUS_HUMAN | CLU | X | X |
| Alpha-2-antiplasmin | P08697 | A2AP_HUMAN | SERPINF2 | X | X |
| Carboxypeptidase N subunit 2 | P22792 | CPN2_HUMAN | CPN2 | X | X |
| Albumin | P02768 | ALBU_HUMAN | ALB | X | X |
| Complement factor B | P00751 | CFAB_HUMAN | CFB | X | X |
| Inter-alpha-trypsin inhibitor heavy chain H4 | Q14624 | ITIH4_HUMAN | ITIH4 | X | X |
| Retinol-binding protein 4 | P02753 | RET4_HUMAN | RBP4 | X | X |
| Complement C1q subcomponent subunit B | P02746 | C1QB_HUMAN | C1QB | X | X |
| Complement C4-B | P0C0L5 | CO4B_HUMAN | C4B | X | X |
| Apolipoprotein A-IV | P06727 | APOA4_HUMAN | APOA4 | X | X |
| Alpha-2-HS-glycoprotein | P02765 | FETUA_HUMAN | AHSG | X | X |
| Beta-2-glycoprotein 1 | P02749 | APOH_HUMAN | APOH | X | X |
| Complement C3 | P01024 | CO3_HUMAN | C3 | X | X |
| Apolipoprotein M | O95445 | APOM_HUMAN | APOM | X | X |
| Protein AMBP | P02760 | AMBP_HUMAN | AMBP | X | X |
| Apolipoprotein B-100 | P04114 | APOB_HUMAN | APOB | X | X |
| Histidine-rich glycoprotein | P04196 | HRG_HUMAN | HRG | X | X |
| Uniprot Code | Gene Name | Protein Name | p-Value | FCh | Response to NAC |
|---|---|---|---|---|---|
| P02741 | CRP | C-reactive protein | 0.00000134 | 6.829624202 | ↓Non-responders |
| P0DOX3 | N/A | Immunoglobulin delta heavy chain | 0.036959856 | 2.75912755 | ↓Non-responders |
| P42858 | HTT | Huntingtin | 0.001165915 | 2.485533233 | ↓Non-responders |
| A0A075B6I1 | IGLV4-60 | Immunoglobulin lambda variable 4-60 | 0.000406597 | 2.458347205 | ↓Non-responders |
| A0A0A0MT36 | IGKV6D-21 | Immunoglobulin kappa variable 6D-21 | 0.003581497 | 2.197533513 | ↓Non-responders |
| P0DJI8 | SAA1 | Serum amyloid A-1 protein | 0.00604557 | 1.859220088 | ↓Non-responders |
| Q15485 | FCN2 | Ficolin-2 | 0.000332323 | 1.677931316 | ↓Non-responders |
| P04211 | IGLV7-43 | Immunoglobulin lambda variable 7-43 | 0.037948547 | 1.658779213 | ↓Non-responders |
| Q08380 | LGALS3BP | Galectin-3-binding protein | 0.006599706 | 1.630292329 | ↓Non-responders |
| P00738 | HP | Haptoglobin | 0.004228108 | 1.588362659 | ↓Non-responders |
| A0A0B4J1V6 | IGHV3-73 | Immunoglobulin heavy variable 3-73 | 0.040907506 | 1.586899833 | ↓Non-responders |
| P0DOX2 | N/A | Immunoglobulin alpha-2 heavy chain | 0.023074607 | 1.573627149 | ↓Non-responders |
| P0C0L5 | C4B | Complement C4-B | 0.015214866 | 1.524647235 | ↓Non-responders |
| P01766 | IGHV3-13 | Immunoglobulin heavy variable 3-13 | 0.021240441 | 1.437890543 | ↓Non-responders |
| P10720 | PF4V1 | Platelet factor 4 variant | 0.016361485 | 1.340407061 | ↓Non-responders |
| P05546 | SERPIND1 | Heparin cofactor 2 | 0.001552533 | 1.328820011 | ↓Non-responders |
| P02743 | APCS | Serum amyloid P-component | 0.019065612 | 1.322698566 | ↓Non-responders |
| P43652 | AFM | Afamin | 0.001267906 | 1.29052425 | ↓Non-responders |
| P02775 | PPBP | Platelet basic protein | 0.016993573 | 1.271442298 | ↓Non-responders |
| P36955 | SERPINF1 | Pigment epithelium-derived factor | 0.000136647 | 1.257360069 | ↓Non-responders |
| P04114 | APOB | Apolipoprotein B-100 | 0.030902399 | 1.257016742 | ↓Non-responders |
| P01009 | SERPINA1 | Alpha-1-antitrypsin | 0.030398754 | 1.252498148 | ↓Non-responders |
| P18428 | LBP | Lipopolysaccharide-binding protein | 0.017544605 | 1.251259211 | ↓Non-responders |
| P25311 | AZGP1 | Zinc-alpha-2-glycoprotein | 0.000669343 | 1.225228845 | ↓Non-responders |
| P02763 | ORM1 | Alpha-1-acid glycoprotein 1 | 0.041098354 | 1.176485834 | ↓Non-responders |
| P02649 | APOE | Apolipoprotein E | 0.048165556 | 1.174660228 | ↓Non-responders |
| P05090 | APOD | Apolipoprotein D | 0.042743468 | 0.873825169 | ↑ Non-responders |
| P22792 | CPN2 | Carboxypeptidase N subunit 2 | 0.029951414 | 0.826212759 | ↑ Non-responders |
| A0A0B4J1X5 | IGHV3-74 | Immunoglobulin heavy variable 3-74 | 0.043884568 | 0.824616939 | ↑ Non-responders |
| P01599 | IGKV1-17 | Immunoglobulin kappa variable 1-17 | 0.014360162 | 0.779005563 | ↑ Non-responders |
| P27169 | PON1 | Serum paraoxonase/arylesterase 1 | 0.001360555 | 0.744636131 | ↑ Non-responders |
| P04433 | IGKV3-11 | Immunoglobulin kappa variable 3-11 | 0.000634898 | 0.712186884 | ↑ Non-responders |
| A0A087WSX0 | IGLV5-45 | Immunoglobulin lambda variable 5-45 | 0.032089451 | 0.703275957 | ↑ Non-responders |
| A0A075B6S5 | IGKV1-27 | Immunoglobulin kappa variable 1-27 | 0.00090962 | 0.698043088 | ↑ Non-responders |
| Q5U7I5 | TTR | Transthyretin | 0.03635507 | 0.684353438 | ↑ Non-responders |
| P01594 | IGKV1-33 | Immunoglobulin kappa variable 1-33 | 0.002337947 | 0.679286204 | ↑ Non-responders |
| A0A0C4DH31 | IGHV1-18 | Immunoglobulin heavy variable 1-18 | 0.006452433 | 0.573322357 | ↑ Non-responders |
| Q9NPH3 | IL1RAP | Interleukin-1 receptor accessory protein | 0.00000586 | 0.518752156 | ↑ Non-responders |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chantada-Vázquez, M.d.P.; Conde-Amboage, M.; Graña-López, L.; Vázquez-Estévez, S.; Bravo, S.B.; Núñez, C. Circulating Proteins Associated with Response and Resistance to Neoadjuvant Chemotherapy in HER2-Positive Breast Cancer. Cancers 2022, 14, 1087. https://doi.org/10.3390/cancers14041087
Chantada-Vázquez MdP, Conde-Amboage M, Graña-López L, Vázquez-Estévez S, Bravo SB, Núñez C. Circulating Proteins Associated with Response and Resistance to Neoadjuvant Chemotherapy in HER2-Positive Breast Cancer. Cancers. 2022; 14(4):1087. https://doi.org/10.3390/cancers14041087
Chicago/Turabian StyleChantada-Vázquez, María del Pilar, Mercedes Conde-Amboage, Lucía Graña-López, Sergio Vázquez-Estévez, Susana B. Bravo, and Cristina Núñez. 2022. "Circulating Proteins Associated with Response and Resistance to Neoadjuvant Chemotherapy in HER2-Positive Breast Cancer" Cancers 14, no. 4: 1087. https://doi.org/10.3390/cancers14041087
APA StyleChantada-Vázquez, M. d. P., Conde-Amboage, M., Graña-López, L., Vázquez-Estévez, S., Bravo, S. B., & Núñez, C. (2022). Circulating Proteins Associated with Response and Resistance to Neoadjuvant Chemotherapy in HER2-Positive Breast Cancer. Cancers, 14(4), 1087. https://doi.org/10.3390/cancers14041087