Identification of Urine Biomarkers to Improve Eligibility for Prostate Biopsy and Detect High-Grade Prostate Cancer
Abstract
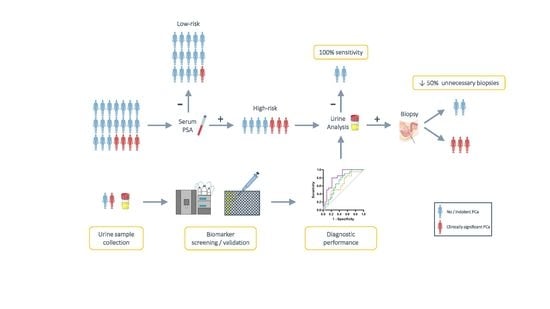
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Urine Collection and Processing
2.2. Mass Spectrometry Analysis
2.2.1. Sample Preparation
2.2.2. Clean-Up for Mass Spectrometry
2.2.3. HPRP Fractionation
2.2.4. Shotgun LC–MS/MS for Spectral Library Generation
2.2.5. HRM Mass Spectrometry Acquisition
2.2.6. Database Search of Shotgun LC–MS/MS Data and Spectral Library Generation
2.2.7. HRM Data Analysis
2.2.8. Data Analysis
2.3. ELISA Validation
2.4. Immunohistochemical Staining of Prostate Tissues
2.5. Statistics and Data Analysis
3. Results
3.1. Patient Characteristics
3.2. Mass Spectrometry Screening and Selection of Urine Biomarkers for PCa Detection
3.3. Increase of PCa Detection Performance through Combinatory Analysis of Biomarkers
3.4. Validation of Biomarker Performance by ELISA
3.5. Immunohistochemical Analysis of Biomarker Expression in Malignant and Healthy Prostate Tissue
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PCa | prostate cancer |
| PSA | prostate specific antigen |
| MS | mass spectrometry |
| AUC | area under the curve |
| MRI | magnetic resonance imaging |
| NMR | nuclear magnetic resonance |
| ELISA | enzyme-linked immunosorbent assay |
| DRE | digital rectal examination |
| HPLC | high-performance liquid chromatography |
| FA | formic acid |
| UV/VIS | ultraviolet–visible |
| HPRP | high-pH reversed-phase chromatography |
| LC–MS | liquid chromatography–mass spectrometry |
| HRM | high resolution mass spectrometry |
| DIA | data-independent acquisition |
| FDR | false discovery rate |
| ROC | receiver operating characteristic |
| HE | Hematoxylin–eosin |
| GS | Gleason score |
| EPV | events per predictor variable |
| PEDF | pigment epithelium-derived factor |
| HPX | hemopexin |
| CD99 | cluster of differentiation 99 |
| CANX | calnexin precursor |
| FCER2 | Fc fragment Of IgE receptor II |
| HRNR | hornerin |
| KRT13 | keratin 13 |
| CD44 | cluster of differentiation 44 |
| RNASE2 | ribonuclease A family member 2 |
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroder, F.H.; Hugosson, J.; Roobol, M.J.; Tammela, T.L.; Zappa, M.; Nelen, V.; Kwiatkowski, M.; Lujan, M.; Maattanen, L.; Lilja, H.; et al. Screening and prostate cancer mortality: Results of the European Randomised Study of Screening for Prostate Cancer (ERSPC) at 13 years of follow-up. Lancet 2014, 384, 2027–2035. [Google Scholar] [CrossRef] [Green Version]
- Osses, D.F.; Roobol, M.J.; Schoots, I.G. Prediction Medicine: Biomarkers, Risk Calculators and Magnetic Resonance Imaging as Risk Stratification Tools in Prostate Cancer Diagnosis. Int. J. Mol. Sci. 2019, 20, 1637. [Google Scholar] [CrossRef] [Green Version]
- Bokhorst, L.P.; Zhu, X.; Bul, M.; Bangma, C.H.; Schroder, F.H.; Roobol, M.J. Positive predictive value of prostate biopsy indicated by prostate-specific-antigen-based prostate cancer screening: Trends over time in a European randomized trial. BJU Int. Br. J. Urol. 2012, 110, 1654–1660. [Google Scholar] [CrossRef] [PubMed]
- Heijnsdijk, E.A.; Wever, E.M.; Auvinen, A.; Hugosson, J.; Ciatto, S.; Nelen, V.; Kwiatkowski, M.; Villers, A.; Paez, A.; Moss, S.M.; et al. Quality-of-life effects of prostate-specific antigen screening. N. Engl. J. Med. 2012, 367, 595–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Poppel, H.; Hogenhout, R.; Albers, P.; van den Bergh, R.C.N.; Barentsz, J.O.; Roobol, M.J. Early Detection of Prostate Cancer in 2020 and Beyond: Facts and Recommendations for the European Union and the European Commission. Eur. Urol. 2021, 79, 327–329. [Google Scholar] [CrossRef] [PubMed]
- Dinges, S.S.; Hohm, A.; Vandergrift, L.A.; Nowak, J.; Habbel, P.; Kaltashov, I.A.; Cheng, L.L. Cancer metabolomic markers in urine: Evidence, techniques and recommendations. Nat. Rev. Urol. 2019, 16, 339–362. [Google Scholar] [CrossRef]
- Tanase, C.P.; Codrici, E.; Popescu, I.D.; Mihai, S.; Enciu, A.M.; Necula, L.G.; Preda, A.; Ismail, G.; Albulescu, R. Prostate cancer proteomics: Current trends and future perspectives for biomarker discovery. Oncotarget 2017, 8, 18497–18512. [Google Scholar] [CrossRef] [Green Version]
- Bax, C.; Lotesoriere, B.J.; Sironi, S.; Capelli, L. Review and Comparison of Cancer Biomarker Trends in Urine as a Basis for New Diagnostic Pathways. Cancers 2019, 11, 1244. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.; Nonomura, N. Urinary biomarkers of prostate cancer. Int. J. Urol. 2018, 25, 770–779. [Google Scholar] [CrossRef] [Green Version]
- Albers, D.D.; Mc, D.J.; Thompson, G.J. Carcinoma cells in prostatic secretions. J. Am. Med. Assoc. 1949, 139, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhao, D.; Spring, D.J.; DePinho, R.A. Genetics and biology of prostate cancer. Genes Dev. 2018, 32, 1105–1140. [Google Scholar] [CrossRef] [Green Version]
- Filella, X.; Foj, L. Prostate Cancer Detection and Prognosis: From Prostate Specific Antigen (PSA) to Exosomal Biomarkers. Int. J. Mol. Sci. 2016, 17, 1784. [Google Scholar] [CrossRef] [PubMed]
- Swensen, A.C.; He, J.; Fang, A.C.; Ye, Y.; Nicora, C.D.; Shi, T.; Liu, A.Y.; Sigdel, T.K.; Sarwal, M.M.; Qian, W.J. A Comprehensive Urine Proteome Database Generated From Patients With Various Renal Conditions and Prostate Cancer. Front. Med. 2021, 8, 548212. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Skotland, T.; Berge, V.; Sandvig, K.; Llorente, A. Exosomal proteins as prostate cancer biomarkers in urine: From mass spectrometry discovery to immunoassay-based validation. Eur. J. Pharm. Sci. 2017, 98, 80–85. [Google Scholar] [CrossRef]
- Taube, S.E.; Clark, G.M.; Dancey, J.E.; McShane, L.M.; Sigman, C.C.; Gutman, S.I. A perspective on challenges and issues in biomarker development and drug and biomarker codevelopment. J. Natl. Cancer Inst. 2009, 101, 1453–1463. [Google Scholar] [CrossRef] [Green Version]
- Fung, K.Y.; Tabor, B.; Buckley, M.J.; Priebe, I.K.; Purins, L.; Pompeia, C.; Brierley, G.V.; Lockett, T.; Gibbs, P.; Tie, J.; et al. Blood-based protein biomarker panel for the detection of colorectal cancer. PLoS ONE 2015, 10, e0120425. [Google Scholar] [CrossRef] [Green Version]
- Kelstrup, C.D.; Young, C.; Lavallee, R.; Nielsen, M.L.; Olsen, J.V. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J. Proteome Res. 2012, 11, 3487–3497. [Google Scholar] [CrossRef]
- Huang, T.; Bruderer, R.; Muntel, J.; Xuan, Y.; Vitek, O.; Reiter, L. Combining Precursor and Fragment Information for Improved Detection of Differential Abundance in Data Independent Acquisition. Mol. Cell Proteom. 2020, 19, 421–430. [Google Scholar] [CrossRef] [Green Version]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [Green Version]
- Loeb, S.; Bjurlin, M.A.; Nicholson, J.; Tammela, T.L.; Penson, D.F.; Carter, H.B.; Carroll, P.; Etzioni, R. Overdiagnosis and overtreatment of prostate cancer. Eur. Urol. 2014, 65, 1046–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jedinak, A.; Loughlin, K.R.; Moses, M.A. Approaches to the discovery of non-invasive urinary biomarkers of prostate cancer. Oncotarget 2018, 9, 32534–32550. [Google Scholar] [CrossRef] [PubMed]
- Khoo, A.; Liu, L.Y.; Nyalwidhe, J.O.; Semmes, O.J.; Vesprini, D.; Downes, M.R.; Boutros, P.C.; Liu, S.K.; Kislinger, T. Proteomic discovery of non-invasive biomarkers of localized prostate cancer using mass spectrometry. Nat. Rev. Urol. 2021, 18, 707–724. [Google Scholar] [CrossRef] [PubMed]
- Gutknecht, M.F.; Seaman, M.E.; Ning, B.; Cornejo, D.A.; Mugler, E.; Antkowiak, P.F.; Moskaluk, C.A.; Hu, S.; Epstein, F.H.; Kelly, K.A. Identification of the S100 fused-type protein hornerin as a regulator of tumor vascularity. Nat. Commun. 2017, 8, 552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.; Kim, D.I.; Kim, J.; Kim, B.H.; Kim, A. Hornerin Is Involved in Breast Cancer Progression. J. Breast Cancer 2016, 19, 142–147. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.J.; Shen, S.L.; Li, S.Q.; Hua, Y.P.; Hu, W.J.; Guo, B.; Peng, B.G. Hornerin promotes tumor progression and is associated with poor prognosis in hepatocellular carcinoma. BMC Cancer 2018, 18, 815. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Hussain, T.; MacLennan, G.T.; Fu, P.; Patel, J.; Mukhtar, H. Differential expression of S100A2 and S100A4 during progression of human prostate adenocarcinoma. J. Clin. Oncol. 2003, 21, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Doll, J.A.; Stellmach, V.M.; Bouck, N.P.; Bergh, A.R.; Lee, C.; Abramson, L.P.; Cornwell, M.L.; Pins, M.R.; Borensztajn, J.; Crawford, S.E. Pigment epithelium-derived factor regulates the vasculature and mass of the prostate and pancreas. Nat. Med. 2003, 9, 774–780. [Google Scholar] [CrossRef]
- Halin, S.; Wikstrom, P.; Rudolfsson, S.H.; Stattin, P.; Doll, J.A.; Crawford, S.E.; Bergh, A. Decreased pigment epithelium-derived factor is associated with metastatic phenotype in human and rat prostate tumors. Cancer Res. 2004, 64, 5664–5671. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Shin, Y.K.; Lee, I.S.; Bae, Y.M.; Sohn, H.W.; Suh, Y.H.; Ree, H.J.; Rowe, M.; Park, S.H. Viral latent membrane protein 1 (LMP-1)-induced CD99 down-regulation in B cells leads to the generation of cells with Hodgkin’s and Reed-Sternberg phenotype. Blood 2000, 95, 294–300. [Google Scholar] [CrossRef]
- Manara, M.C.; Bernard, G.; Lollini, P.L.; Nanni, P.; Zuntini, M.; Landuzzi, L.; Benini, S.; Lattanzi, G.; Sciandra, M.; Serra, M.; et al. CD99 acts as an oncosuppressor in osteosarcoma. Mol. Biol. Cell 2006, 17, 1910–1921. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.C.; Park, W.S.; Bae, Y.M.; Hahn, J.H.; Hahn, K.; Lee, H.; Lee, H.W.; Koo, H.J.; Shin, H.J.; Shin, H.S.; et al. Immunoreactivity of CD99 in stomach cancer. J. Korean Med. Sci. 2002, 17, 483–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scotlandi, K.; Zuntini, M.; Manara, M.C.; Sciandra, M.; Rocchi, A.; Benini, S.; Nicoletti, G.; Bernard, G.; Nanni, P.; Lollini, P.L.; et al. CD99 isoforms dictate opposite functions in tumour malignancy and metastases by activating or repressing c-Src kinase activity. Oncogene 2007, 26, 6604–6618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davalieva, K.; Kiprijanovska, S.; Maleva Kostovska, I.; Stavridis, S.; Stankov, O.; Komina, S.; Petrusevska, G.; Polenakovic, M. Comparative Proteomics Analysis of Urine Reveals Down-Regulation of Acute Phase Response Signaling and LXR/RXR Activation Pathways in Prostate Cancer. Proteomes 2018, 6, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lima, T.; Henrique, R.; Vitorino, R.; Fardilha, M. Bioinformatic analysis of dysregulated proteins in prostate cancer patients reveals putative urinary biomarkers and key biological pathways. Med. Oncol. 2021, 38, 9. [Google Scholar] [CrossRef]
- Sarfati, M.; Bron, D.; Lagneaux, L.; Fonteyn, C.; Frost, H.; Delespesse, G. Elevation of IgE-binding factors in serum of patients with B cell-derived chronic lymphocytic leukemia. Blood 1988, 71, 94–98. [Google Scholar] [CrossRef]
- Caligaris-Cappio, F.; Ghia, P. The normal counterpart to the chronic lymphocytic leukemia B cell. Best Pract. Res. Clin. Haematol. 2007, 20, 385–397. [Google Scholar] [CrossRef]
- Barna, G.; Reiniger, L.; Tatrai, P.; Kopper, L.; Matolcsy, A. The cut-off levels of CD23 expression in the differential diagnosis of MCL and CLL. Hematol. Oncol. 2008, 26, 167–170. [Google Scholar] [CrossRef]
- Schlette, E.; Fu, K.; Medeiros, L.J. CD23 expression in mantle cell lymphoma: Clinicopathologic features of 18 cases. Am. J. Clin. Pathol. 2003, 120, 760–766. [Google Scholar] [CrossRef]
- Walters, M.; Olteanu, H.; Van Tuinen, P.; Kroft, S.H. CD23 expression in plasma cell myeloma is specific for abnormalities of chromosome 11, and is associated with primary plasma cell leukaemia in this cytogenetic sub-group. Br. J. Haematol. 2010, 149, 292–293. [Google Scholar] [CrossRef]
- Soriano, A.O.; Thompson, M.A.; Admirand, J.H.; Fayad, L.E.; Rodriguez, A.M.; Romaguera, J.E.; Hagemeister, F.B.; Pro, B. Follicular dendritic cell sarcoma: A report of 14 cases and a review of the literature. Am. J. Hematol. 2007, 82, 725–728. [Google Scholar] [CrossRef] [PubMed]
- Peter Rieber, E.; Rank, G.; Köhler, I.; Krauss, S. Membrane Expression of Fc∈RII/CD23 and Release of Soluble CD23 by Follicular Dendritic Cells. In Dendritic Cells in Fundamental and Clinical Immunology; Kamperdijk, E.W.A., Nieuwenhuis, P., Hoefsmit, E.C.M., Eds.; Springer: New York, NY, USA, 1993; pp. 393–398. [Google Scholar]
- Ida-Yonemochi, H.; Maruyama, S.; Kobayashi, T.; Yamazaki, M.; Cheng, J.; Saku, T. Loss of keratin 13 in oral carcinoma in situ: A comparative study of protein and gene expression levels using paraffin sections. Mod. Pathol. 2012, 25, 784–794. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, K.; Aragaki, T.; Morita, K.; Kawachi, H.; Kayamori, K.; Nakanishi, S.; Omura, K.; Miki, Y.; Okada, N.; Katsube, K.; et al. Down-regulation of keratin 4 and keratin 13 expression in oral squamous cell carcinoma and epithelial dysplasia: A clue for histopathogenesis. Histopathology 2011, 58, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, K.; Hatta, M.; Ikebe, T.; Yamazaki, J. Epigenetic alterations of the keratin 13 gene in oral squamous cell carcinoma. BMC Cancer 2014, 14, 988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsit, C.J.; Houseman, E.A.; Christensen, B.C.; Gagne, L.; Wrensch, M.R.; Nelson, H.H.; Wiemels, J.; Zheng, S.; Wiencke, J.K.; Andrew, A.S.; et al. Identification of methylated genes associated with aggressive bladder cancer. PLoS ONE 2010, 5, e12334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Yin, L.; Jones, L.W.; Chu, G.C.; Wu, J.B.; Huang, J.M.; Li, Q.; You, S.; Kim, J.; Lu, Y.T.; et al. Keratin 13 expression reprograms bone and brain metastases of human prostate cancer cells. Oncotarget 2016, 7, 84645–84657. [Google Scholar] [CrossRef] [PubMed]
- Rüschoff, J.H.; Stratton, S.; Roberts, E.; Clark, S.; Sebastiao, N.; Fankhauser, C.D.; Eberli, D.; Moch, H.; Wild, P.J.; Rupp, N.J. A novel 5x multiplex immunohistochemical staining reveals PSMA as a helpful marker in prostate cancer with low p504s expression. Pathol. Res. Pract. 2021, 228, 153667. [Google Scholar] [CrossRef]
- Schrag, J.D.; Bergeron, J.J.; Li, Y.; Borisova, S.; Hahn, M.; Thomas, D.Y.; Cygler, M. The Structure of calnexin, an ER chaperone involved in quality control of protein folding. Mol. Cell 2001, 8, 633–644. [Google Scholar] [CrossRef]
- Dissemond, J.; Busch, M.; Kothen, T.; Mörs, J.; Weimann, T.K.; Lindeke, A.; Goos, M.; Wagner, S.N. Differential downregulation of endoplasmic reticulum-residing chaperones calnexin and calreticulin in human metastatic melanoma. Cancer Lett. 2004, 203, 225–231. [Google Scholar] [CrossRef]
- Ryan, D.; Carberry, S.; Murphy, Á.C.; Lindner, A.U.; Fay, J.; Hector, S.; McCawley, N.; Bacon, O.; Concannon, C.G.; Kay, E.W.; et al. Calnexin, an ER-induced protein, is a prognostic marker and potential therapeutic target in colorectal cancer. J. Transl. Med. 2016, 14, 196. [Google Scholar] [CrossRef] [Green Version]
- Hibbert, R.G.; Teriete, P.; Grundy, G.J.; Beavil, R.L.; Reljic, R.; Holers, V.M.; Hannan, J.P.; Sutton, B.J.; Gould, H.J.; McDonnell, J.M. The structure of human CD23 and its interactions with IgE and CD21. J. Exp. Med. 2005, 202, 751–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vittinghoff, E.; McCulloch, C.E. Relaxing the Rule of Ten Events per Variable in Logistic and Cox Regression. Am. J. Epidemiol. 2006, 165, 710–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Number of Samples (% of Total) | Gleason Score | Median Age (Min–Max) | Median Serum PSA (Min–Max) | Prostate Volume (Min–Max) * | |
|---|---|---|---|---|---|
| No Tumor | 24 (53.3%) | 0 | 63.5 (52–82) | 6.60 (2.00–14.97) | 60.19 (18.56–203.68) |
| Tumor | 21 (46.7%) | 6–9 | 65 (52–76) | 7.22 (2.00–38.80) | 48.59 (17.00–80.63) |
| 4 (8.9%) | 6 | 65 (64–70) | 8.53 (4.53–17.37) | 60.54 (30.90–80.63) | |
| 8 (17.8%) | 7 | 65 (52–73) | 4.94 (2.00–11.00) | 50.00 (26.45–72.54) | |
| 9 (20.0%) | 8–9 | 74 (58–76) | 12.41 (4.86–38.80) | 47.17 (17.00–60.00) | |
| Total | 45 (100%) | 65 (52–82) | 6.90 (2.00–38.80) | 52.00 (17.00–203.68) |
| Biomarker | AUC | Std. Error | 95% Confidence Interval | p-Value | Specificity at 90% Sensitivity | Specificity at 100% Sensitivity |
|---|---|---|---|---|---|---|
| PEDF | 0.8023 | 0.070 | 0.6659 to 0.9386 | 0.0008 | 68.2 | 36.4 |
| HPX | 0.7761 | 0.070 | 0.6396 to 0.9125 | 0.0020 | 52.2 | 39.1 |
| HRNR | 0.7522 | 0.076 | 0.6033 to 0.9010 | 0.0047 | 47.8 | 13.0 |
| KRT13 | 0.7391 | 0.075 | 0.5913 to 0.8869 | 0.0074 | 52.2 | 30.4 |
| CANX | 0.7043 | 0.085 | 0.5377 to 0.8708 | 0.0273 | 47.6 | 38.1 |
| CD99 | 0.6750 | 0.083 | 0.5114 to 0.8386 | 0.0525 | 36.4 | 31.8 |
| FCER2 | 0.6717 | 0.084 | 0.5075 to 0.8360 | 0.0544 | 52.2 | 30.4 |
| PEDF + HPX | 0.8977 | 0.050 | 0.7999 to 0.9956 | <0.0001 | 72.7 | 50.0 |
| PEDF + CD99 | 0.8786 | 0.056 | 0.7689 to 0.9883 | <0.0001 | 76.2 | 66.7 |
| PEDF + FCER2 | 0.8773 | 0.063 | 0.7530 to 1.000 | <0.0001 | 86.4 | 72.7 |
| PEDF + KRT13 | 0.8705 | 0.055 | 0.7618 to 0.9791 | <0.0001 | 72.7 | 54.5 |
| PEDF + HRNR | 0.8568 | 0.058 | 0.7437 to 0.9699 | <0.0001 | 77.3 | 54.5 |
| PEDF + CANX | 0.9105 | 0.053 | 0.8067 to 1.000 | <0.0001 | 85.0 | 70.0 |
| HPX + HRNR | 0.8739 | 0.054 | 0.7682 to 0.9797 | <0.0001 | 73.9 | 34.8 |
| HPX + KRT13 | 0.8413 | 0.061 | 0.7211 to 0.9615 | 0.0001 | 60.9 | 56.5 |
| HRNR + CANX | 0.8496 | 0.062 | 0.7272 to 0.9720 | 0.0002 | 66.7 | 66.7 |
| HPX + FCER2 | 0.8000 | 0.068 | 0.6670 to 0.9330 | 0.0008 | 60.9 | 60.9 |
| HPX + CD99 | 0.7864 | 0.071 | 0.6462 to 0.9265 | 0.0015 | 63.6 | 54.5 |
| KRT13 + CANX | 0.7820 | 0.076 | 0.6322 to 0.9318 | 0.0023 | 61.9 | 61.9 |
| KRT13 + FCER2 | 0.7652 | 0.074 | 0.6193 to 0.9111 | 0.0030 | 60.9 | 47.8 |
| HRNR + FCER2 | 0.7457 | 0.076 | 0.5964 to 0.8949 | 0.0059 | 60.9 | 34.8 |
| Biomarker | AUC | Std. Error | 95% Confidence Interval | p-Value | Specificity at 90% Sensitivity | Specificity at 100% Sensitivity | |
|---|---|---|---|---|---|---|---|
| All PCa grades | KRT13 | 0.8087 | 0.066 | 0.6797 to 0.9377 | 0.0005 | 43.5 | 43.5 |
| HPX | 0.7696 | 0.071 | 0.6314 to 0.9077 | 0.0025 | 47.8 | 43.5 | |
| PEDF | 0.7609 | 0.073 | 0.6176 to 0.9041 | 0.0035 | 34.8 | 30.4 | |
| CD99 | 0.7565 | 0.073 | 0.6136 to 0.8994 | 0.0041 | 52.2 | 47.8 | |
| FCER2 | 0.7565 | 0.074 | 0.6114 to 0.9017 | 0.0041 | 47.8 | 13.0 | |
| CANX | 0.7457 | 0.076 | 0.5971 to 0.8942 | 0.0059 | 30.4 | 26.1 | |
| HRNR | 0.7120 | 0.080 | 0.5553 to 0.8686 | 0.0176 | 39.1 | 17.4 | |
| High-grade PCa | KRT13 | 0.7708 | 0.075 | 0.6247 to 0.9170 | 0.0033 | 40.7 | 37.1 |
| HPX | 0.7546 | 0.074 | 0.6094 to 0.8998 | 0.0057 | 44.4 | 37.0 | |
| PEDF | 0.7292 | 0.079 | 0.5752 to 0.8831 | 0.0129 | 33.3 | 29.6 | |
| FCER2 | 0.7269 | 0.081 | 0.5690 to 0.8847 | 0.0138 | 44.4 | 11.2 | |
| CD99 | 0.7222 | 0.078 | 0.5688 to 0.8756 | 0.0159 | 40.7 | 40.7 | |
| HRNR | 0.6956 | 0.083 | 0.5321 to 0.8591 | 0.0337 | 37.0 | 14.8 | |
| CANX | 0.6528 | 0.086 | 0.4849 to 0.8207 | 0.0973 | 25.9 | 22.1 |
| Biomarker | AUC | Std. Error | 95% Confidence Interval | p-Value | Specificity at 90% Sensitivity | Specificity at 100% Sensitivity | |
|---|---|---|---|---|---|---|---|
| All PCa grades | KRT13 | 0.7696 | 0.071 | 0.6298 to 0.9093 | 0.0025 | 52.2 | 30.4 |
| HRNR | 0.7413 | 0.079 | 0.5865 to 0.8961 | 0.0069 | 52.2 | 8.7 | |
| FCER2 | 0.7326 | 0.077 | 0.5813 to 0.8839 | 0.0092 | 52.2 | 39.1 | |
| CANX | 0.7043 | 0.080 | 0.5479 to 0.8608 | 0.0221 | 30.4 | 17.4 | |
| PEDF | 0.700 | 0.081 | 0.5404 to 0.8596 | 0.0251 | 30.4 | 30.4 | |
| HPX | 0.6978 | 0.081 | 0.5386 to 0.8570 | 0.0267 | 39.1 | 8.7 | |
| CD99 | 0.6652 | 0.083 | 0.5032 to 0.8273 | 0.0642 | 34.8 | 21.7 | |
| KRT13 + FCER2 | 0.8196 | 0.065 | 0.6927 to 0.9464 | 0.0003 | 52.2 | 52.2 | |
| HPX + FCER2 | 0.8087 | 0.067 | 0.6767 to 0.9407 | 0.0005 | 43.5 | 30.4 | |
| PEDF + FCER2 | 0.8022 | 0.067 | 0.6714 to 0.9329 | 0.0007 | 52.2 | 39.1 | |
| HPX + KRT13 | 0.7826 | 0.070 | 0.6462 to 0.9190 | 0.0015 | 52.2 | 30.4 | |
| HRNR + FCER2 | 0.7826 | 0.071 | 0.6429 to 0.9223 | 0.0015 | 56.5 | 13.0 | |
| PEDF + KRT13 | 0.7804 | 0.070 | 0.6431 to 0.9178 | 0.0017 | 52.2 | 39.1 | |
| KRT13 + CANX | 0.7609 | 0.072 | 0.6189 to 0.9028 | 0.0035 | 47.8 | 30.4 | |
| HPX + HRNR | 0.7478 | 0.078 | 0.5960 to 0.8997 | 0.0055 | 43.5 | 8.7 | |
| PEDF + CANX | 0.7348 | 0.077 | 0.5844 to 0.8852 | 0.0085 | 47.8 | 26.1 | |
| HRNR + CANX | 0.7326 | 0.079 | 0.5781 to 0.8871 | 0.0092 | 43.5 | 8.7 | |
| PEDF + CD99 | 0.7304 | 0.076 | 0.5808 to 0.8801 | 0.0099 | 43.5 | 34.8 | |
| PEDF + HRNR | 0.7283 | 0.080 | 0.5723 to 0.8842 | 0.0106 | 43.5 | 8.7 | |
| HPX + CD99 | 0.7283 | 0.078 | 0.5753 to 0.8812 | 0.0106 | 39.1 | 17.4 | |
| PEDF + HPX | 0.7000 | 0.081 | 0.5417 to 0.8583 | 0.0251 | 26.1 | 13.0 | |
| High-grade PCa | KRT13 | 0.7361 | 0.077 | 0.5854 to 0.8868 | 0.0104 | 40.7 | 25.9 |
| HRNR | 0.7199 | 0.084 | 0.5551 to 0.8847 | 0.0170 | 14.8 | 7.4 | |
| FCER2 | 0.7014 | 0.079 | 0.5468 to 0.8560 | 0.0288 | 44.4 | 33.3 | |
| HPX | 0.6968 | 0.087 | 0.5262 to 0.8673 | 0.0327 | 7.4 | 7.4 | |
| PEDF | 0.6806 | 0.085 | 0.5141 to 0.8470 | 0.0500 | 33.3 | 18.5 | |
| CD99 | 0.6644 | 0.086 | 0.4967 to 0.8320 | 0.0744 | 29.6 | 18.5 | |
| CANX | 0.6574 | 0.085 | 0.4907 to 0.8241 | 0.0875 | 22.2 | 14.8 | |
| HPX + FCER2 | 0.7894 | 0.077 | 0.6376 to 0.9411 | 0.0017 | 33.3 | 33.3 | |
| HPX + KRT13 | 0.7870 | 0.073 | 0.6432 to 0.9308 | 0.0018 | 33.3 | 18.5 | |
| KRT13 + FCER2 | 0.7801 | 0.069 | 0.6447 to 0.9155 | 0.0024 | 51.8 | 48.1 | |
| HPX + CD99 | 0.7662 | 0.078 | 0.6136 to 0.9188 | 0.0039 | 29.6 | 14.8 | |
| PEDF + FCER2 | 0.7523 | 0.073 | 0.6090 to 0.8956 | 0.0062 | 48.1 | 44.5 | |
| HRNR + FCER2 | 0.7523 | 0.076 | 0.6024 to 0.9022 | 0.0062 | 51.8 | 11.1 | |
| HPX + HRNR | 0.7500 | 0.084 | 0.5845 to 0.9155 | 0.0067 | 11.1 | 7.4 | |
| PEDF + KRT13 | 0.7431 | 0.075 | 0.5964 to 0.8898 | 0.0083 | 44.5 | 33.3 | |
| KRT13 + CANX | 0.7384 | 0.076 | 0.5886 to 0.8882 | 0.0097 | 40.7 | 29.6 | |
| PEDF + CD99 | 0.7176 | 0.078 | 0.5657 to 0.8695 | 0.0182 | 37.0 | 37.0 | |
| PEDF + HPX | 0.7083 | 0.083 | 0.5461 to 0.8705 | 0.0237 | 14.8 | 14.8 | |
| HRNR + CANX | 0.7014 | 0.083 | 0.5384 to 0.8644 | 0.0288 | 29.6 | 3.7 | |
| PEDF + HRNR | 0.6968 | 0.082 | 0.5358 to 0.8577 | 0.0327 | 33.3 | 11.1 | |
| PEDF + CANX | 0.6898 | 0.081 | 0.5303 to 0.8493 | 0.0394 | 44.4 | 18.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alijaj, N.; Pavlovic, B.; Martel, P.; Rakauskas, A.; Cesson, V.; Saba, K.; Hermanns, T.; Oechslin, P.; Veit, M.; Provenzano, M.; et al. Identification of Urine Biomarkers to Improve Eligibility for Prostate Biopsy and Detect High-Grade Prostate Cancer. Cancers 2022, 14, 1135. https://doi.org/10.3390/cancers14051135
Alijaj N, Pavlovic B, Martel P, Rakauskas A, Cesson V, Saba K, Hermanns T, Oechslin P, Veit M, Provenzano M, et al. Identification of Urine Biomarkers to Improve Eligibility for Prostate Biopsy and Detect High-Grade Prostate Cancer. Cancers. 2022; 14(5):1135. https://doi.org/10.3390/cancers14051135
Chicago/Turabian StyleAlijaj, Nagjie, Blaz Pavlovic, Paul Martel, Arnas Rakauskas, Valérie Cesson, Karim Saba, Thomas Hermanns, Pascal Oechslin, Markus Veit, Maurizio Provenzano, and et al. 2022. "Identification of Urine Biomarkers to Improve Eligibility for Prostate Biopsy and Detect High-Grade Prostate Cancer" Cancers 14, no. 5: 1135. https://doi.org/10.3390/cancers14051135
APA StyleAlijaj, N., Pavlovic, B., Martel, P., Rakauskas, A., Cesson, V., Saba, K., Hermanns, T., Oechslin, P., Veit, M., Provenzano, M., Rüschoff, J. H., Brada, M. D., Rupp, N. J., Poyet, C., Derré, L., Valerio, M., Banzola, I., & Eberli, D. (2022). Identification of Urine Biomarkers to Improve Eligibility for Prostate Biopsy and Detect High-Grade Prostate Cancer. Cancers, 14(5), 1135. https://doi.org/10.3390/cancers14051135