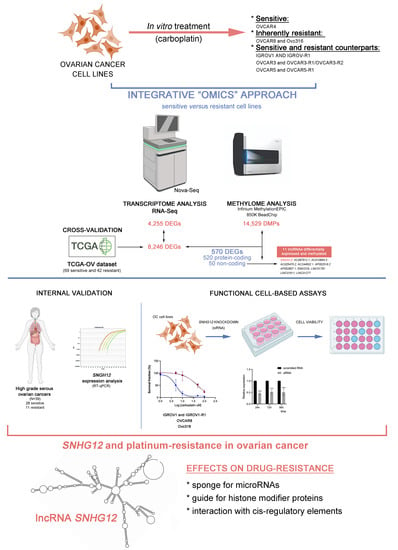
The Long Non-Coding RNA SNHG12 as a Mediator of Carboplatin Resistance in Ovarian Cancer via Epigenetic Mechanisms
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Patient Samples
2.2. Establishment of Resistant Cell Lines
2.3. RNA Extraction and Sequencing
2.4. Pathway Enrichment Analysis
2.5. DNA Extraction and DNA Methylation Profiling
2.6. RNA-seq and DNA Methylation Integrative Analysis
2.7. External Data from The Cancer Genome Atlas
2.8. Criteria Used to Select lncRNA for Functional Assays
2.9. RNA Extraction and RT-qPCR Assay
2.10. Knockdown of lncRNA SNHG12 and Carboplatin Exposure
2.11. Cell Viability Assays
2.12. Data Processing and Statistical Analyses
3. Results
3.1. Carboplatin Response in Ovarian Cancer Cell Lines and Establishment of Carboplatin-Resistant Derived Subpopulations
3.2. Differential Expression Profiles Associated with Carboplatin Resistance in Ovarian Cancer Cell Lines and Ovarian Cancer Tissues
3.3. Differential Methylation Profile Associated with Carboplatin Resistance in Ovarian Cancer Cell Lines
3.4. Integrative Data Analysis
3.5. Platinum Resistance-Associated Genes
3.6. The lncRNA SNHG12 Is Overexpressed in Ovarian Cancer
3.7. Functional Analysis of the lncRNA SNHG12
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian Cancer. Nat. Rev. Dis. Prim. 2016, 2, 16061. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.K.; Bashashati, A.; Anglesio, M.S.; Cochrane, D.R.; Grewal, D.S.; Ha, G.; McPherson, A.; Horlings, H.M.; Senz, J.; Prentice, L.M.; et al. Genomic Consequences of Aberrant DNA Repair Mechanisms Stratify Ovarian Cancer Histotypes. Nat. Genet. 2017, 49, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Lheureux, S.; Gourley, C.; Vergote, I.; Oza, A.M. Epithelial Ovarian Cancer. Lancet 2019, 393, 1240–1253. [Google Scholar] [CrossRef] [Green Version]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Dyba, T.; Randi, G.; Bettio, M.; Gavin, A.; Visser, O.; Bray, F. Cancer Incidence and Mortality Patterns in Europe: Estimates for 40 Countries and 25 Major Cancers in 2018. Eur. J. Cancer 2018, 103, 356–387. [Google Scholar] [CrossRef]
- Boyd, L.R.; Muggia, F.M. Carboplatin/Paclitaxel Induction in Ovarian Cancer: The Finer Points. Oncology 2018, 32, 422–424. [Google Scholar]
- Hennessy, B.T.; Coleman, R.L.; Markman, M. Ovarian Cancer. Lancet 2009, 374, 1371–1382. [Google Scholar] [CrossRef]
- Colombo, N.; Sessa, C.; du Bois, A.; Ledermann, J.; McCluggage, W.G.; McNeish, I.; Morice, P.; Pignata, S.; Ray-Coquard, I.; Vergote, I.; et al. ESMO–ESGO Consensus Conference Recommendations on Ovarian Cancer: Pathology and Molecular Biology, Early and Advanced Stages, Borderline Tumours and Recurrent Disease. Ann. Oncol. 2019, 30, 672–705. [Google Scholar] [CrossRef] [Green Version]
- Jayson, G.C.; Kohn, E.C.; Kitchener, H.C.; Ledermann, J.A. Ovarian Cancer. Lancet 2014, 384, 1376–1388. [Google Scholar] [CrossRef]
- Patch, A.M.; Christie, E.L.; Etemadmoghadam, D.; Garsed, D.W.; George, J.; Fereday, S.; Nones, K.; Cowin, P.; Alsop, K.; Bailey, P.J.; et al. Whole-Genome Characterization of Chemoresistant Ovarian Cancer. Nature 2015, 521, 489–494. [Google Scholar] [CrossRef]
- Danske Gynækologisk Cancer Gruppe. Dansk Gynækologisk Cancer Database—National Årsrapport 2015–2016. Regionernes Kliniske Kvalitetsudviklingsprogram. Available online: www.rkkp.dk (accessed on 22 March 2022).
- Kurnit, K.C.; Fleming, G.F.; Lengyel, E. Updates and New Options in Advanced Epithelial Ovarian Cancer Treatment. Obstet. Gynecol. 2021, 137, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Tew, W.P.; Lacchetti, C.; Ellis, A.; Maxian, K.; Banerjee, S.; Bookman, M.; Jones, M.B.; Lee, J.-M.; Lheureux, S.; Liu, J.F.; et al. PARP Inhibitors in the Management of Ovarian Cancer: ASCO Guideline. J. Clin. Oncol. 2020, 38, 3468–3493. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.K.; Alvarez, R.D.; Bakkum-Gamez, J.N.; Barroilhet, L.; Behbakht, K.; Berchuck, A.; Chen, L.; Cristea, M.; DeRosa, M.; Eisenhauer, E.L.; et al. Ovarian Cancer, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 191–226. [Google Scholar] [CrossRef]
- Fidler, I.J.; Kripke, M.L. The Challenge of Targeting Metastasis. Cancer Metastasis Rev. 2015, 34, 635–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashashati, A.; Ha, G.; Tone, A.; Ding, J.; Prentice, L.M.; Roth, A.; Rosner, J.; Shumansky, K.; Kalloger, S.; Senz, J.; et al. Distinct Evolutionary Trajectories of Primary High-grade Serous Ovarian Cancers Revealed through Spatial Mutational Profiling. J. Pathol. 2013, 231, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Abildgaard, C.; Do Canto, L.M.; Steffensen, K.D.; Rogatto, S.R. Long Non-Coding RNAs Involved in Resistance to Chemotherapy in Ovarian Cancer. Front. Oncol. 2020, 9, 1549. [Google Scholar] [CrossRef] [Green Version]
- Fang, F.; Cardenas, H.; Huang, H.; Jiang, G.; Perkins, S.M.; Zhang, C.; Keer, H.N.; Liu, Y.; Nephew, K.P.; Matei, D. Genomic and Epigenomic Signatures in Ovarian Cancer Associated with Resensitization to Platinum Drugs. Cancer Res. 2018, 78, 631–644. [Google Scholar] [CrossRef] [Green Version]
- Guttman, M.; Rinn, J.L. Modular Regulatory Principles of Large Non-Coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef] [Green Version]
- Bouckenheimer, J.; Assou, S.; Riquier, S.; Hou, C.; Philippe, N.; Sansac, C.; Lavabre-Bertrand, T.; Commes, T.; Lemaître, J.-M.; Boureux, A.; et al. Long Non-Coding RNAs in Human Early Embryonic Development and Their Potential in ART. Hum. Reprod. Update 2016, 23, 19–40. [Google Scholar] [CrossRef]
- Lv, W.; Jia, Y.; Wang, J.; Duan, Y.; Wang, X.; Liu, T.; Hao, S.; Liu, L. Long Non-Coding RNA SNHG10 Upregulates BIN1 to Suppress the Tumorigenesis and Epithelial–Mesenchymal Transition of Epithelial Ovarian Cancer via Sponging MiR-200a-3p. Cell Death Discov. 2022, 8, 60. [Google Scholar] [CrossRef]
- Qiu, J.; Lin, Y.; Ye, L.; Ding, J.; Feng, W.; Jin, H.; Zhang, Y.; Li, Q.; Hua, K. Overexpression of Long Non-Coding RNA HOTAIR Predicts Poor Patient Prognosis and Promotes Tumor Metastasis in Epithelial Ovarian Cancer. Gynecol. Oncol. 2014, 134, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Nikpayam, E.; Tasharrofi, B.; Sarrafzadeh, S.; Ghafouri-Fard, S. The Role of Long Non-Coding RNAs in Ovarian Cancer. Iran. Biomed. J. 2017, 21, 3–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Han, L.; Zhou, L.; Wang, L.; Zhang, L. Prediction of Candidate RNA Signatures for Recurrent Ovarian Cancer Prognosis by the Construction of an Integrated Competing Endogenous RNA Network. Oncol. Rep. 2018, 40, 2659–2673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Zhang, W.; Wang, S.; Liu, K.; Song, F.; Ran, L. A Panel of 7 Prognosis-Related Long Non-Coding RNAs to Improve Platinum-Based Chemoresistance Prediction in Ovarian Cancer. Int. J. Oncol. 2018, 53, 866–876. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhao, Y.; Dong, S.; Liu, L.; Tai, L.; Xu, Y. Systematic Identification of Dysregulated LncRNAs Associated with Platinum-Based Chemotherapy Response across 11 Cancer Types. Genomics 2020, 112, 1214–1222. [Google Scholar] [CrossRef]
- Shi, C.; Wang, M. LINC01118 Modulates Paclitaxel Resistance of Epithelial Ovarian Cancer by Regulating MiR-134/ABCC1. Med. Sci. Monit. 2018, 24, 8831–8839. [Google Scholar] [CrossRef]
- Wang, J.; Ye, C.; Liu, J.; Hu, Y. UCA1 Confers Paclitaxel Resistance to Ovarian Cancer through MiR-129/ABCB1 Axis. Biochem. Biophys. Res. Commun. 2018, 501, 1034–1040. [Google Scholar] [CrossRef]
- Liu, S.; Zou, B.; Tian, T.; Luo, X.; Mao, B.; Zhang, X.; Lei, H. Overexpression of the LncRNA FER1L4 Inhibits Paclitaxel Tolerance of Ovarian Cancer Cells via the Regulation of the MAPK Signaling Pathway. J. Cell. Biochem. 2019, 120, 7581–7589. [Google Scholar] [CrossRef]
- Slaby, O.; Laga, R.; Sedlacek, O. Therapeutic Targeting of Non-Coding RNAs in Cancer. Biochem. J. 2017, 474, 4219–4251. [Google Scholar] [CrossRef]
- Tumours of the Ovary. WHO Classification of Tumours: Female Genital Tumours; WHO Classification of Tumours Editorial Board, Ed.; IARC: Lyon, France, 2020. [Google Scholar]
- Abildgaard, C.; Dahl, C.; Abdul-Al, A.; Christensen, A.; Guldberg, P. Inhibition of Retinoic Acid Receptor β Signaling Confers Glycolytic Dependence and Sensitization to Dichloroacetate in Melanoma Cells. Oncotarget 2017, 8, 84210–84223. [Google Scholar] [CrossRef] [Green Version]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal Violet Assay for Determining Viability of Cultured Cells. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A Tool for Multi-Genome Mapping and Quality Control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- do Canto, L.M.; Barros-Filho, M.C.; Rainho, C.A.; Marinho, D.; Kupper, B.E.C.; de Souza Begnami, M.D.F.; Scapulatempo-Neto, C.; Havelund, B.M.; Lindebjerg, J.; Marchi, F.A.; et al. Comprehensive Analysis of DNA Methylation and Prediction of Response to NeoadjuvantTherapy in Locally Advanced Rectal Cancer. Cancers 2020, 12, 3079. [Google Scholar] [CrossRef]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A Flexible and Comprehensive Bioconductor Package for the Analysis of Infinium DNA Methylation Microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [Green Version]
- Teschendorff, A.E.; Marabita, F.; Lechner, M.; Bartlett, T.; Tegner, J.; Gomez-Cabrero, D.; Beck, S. A Beta-Mixture Quantile Normalization Method for Correcting Probe Design Bias in Illumina Infinium 450 k DNA Methylation Data. Bioinformatics 2013, 29, 189–196. [Google Scholar] [CrossRef] [Green Version]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The Sva Package for Removing Batch Effects and Other Unwanted Variation in High-Throughput Experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Pidsley, R.; Zotenko, E.; Peters, T.J.; Lawrence, M.G.; Risbridger, G.P.; Molloy, P.; Van Djik, S.; Muhlhausler, B.; Stirzaker, C.; Clark, S.J. Critical Evaluation of the Illumina MethylationEPIC BeadChip Microarray for Whole-Genome DNA Methylation Profiling. Genome Biol. 2016, 17, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Liu, R.; Zeng, Y.; Zhou, C.-F.; Wang, Y.; Li, X.; Liu, Z.-Q.; Chen, X.-P.; Zhang, W.; Zhou, H.-H. Long Noncoding RNA Expression Signature to Predict Platinum-Based Chemotherapeutic Sensitivity of Ovarian Cancer Patients. Sci. Rep. 2017, 7, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Guo, Q.; Qi, Y.; Hao, Y.; Gao, Y.; Zhi, H.; Zhang, Y.; Sun, Y.; Zhang, Y.; Xin, M.; et al. LncACTdb 3.0: An Updated Database of Experimentally Supported CeRNA Interactions and Personalized Networks Contributing to Precision Medicine. Nucleic Acids Res. 2022, 50, D183–D189. [Google Scholar] [CrossRef]
- Bao, Z.; Yang, Z.; Huang, Z.; Zhou, Y.; Cui, Q.; Dong, D. LncRNADisease 2.0: An Updated Database of Long Non-Coding RNA-Associated Diseases. Nucleic Acids Res. 2019, 47, D1034–D1037. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Shang, S.; Guo, S.; Li, X.; Zhou, H.; Liu, H.; Sun, Y.; Wang, J.; Wang, P.; Zhi, H.; et al. Lnc2Cancer 3.0: An Updated Resource for Experimentally Supported LncRNA/CircRNA Cancer Associations and Web Tools Based on RNA-Seq and ScRNA-Seq Data. Nucleic Acids Res. 2021, 49, D1251–D1258. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Huang, D.; Savage, S.R.; Calinawan, A.P.; Lin, C.; Zhang, B.; Wang, P.; Starr, T.K.; Birrer, M.J.; Paulovich, A.G. A Highly Annotated Database of Genes Associated with Platinum Resistance in Cancer. Oncogene 2021, 40, 6395–6405. [Google Scholar] [CrossRef]
- Bell, D.; Berchuck, A.; Birrer, M.; Chien, J.; Cramer, D.W.; Dao, F.; Dhir, R.; Disaia, P.; Gabra, H.; Glenn, P.; et al. Integrated Genomic Analyses of Ovarian Carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Lee, S.; Zhao, L.; Rojas, C.; Bateman, N.W.; Yao, H.; Lara, O.D.; Celestino, J.; Morgan, M.B.; Nguyen, T.V.; Conrads, K.A.; et al. Molecular Analysis of Clinically Defined Subsets of High-Grade Serous Ovarian Cancer. Cell Rep. 2020, 31, 107502. [Google Scholar] [CrossRef] [PubMed]
- Talhouk, A.; George, J.; Wang, C.; Budden, T.; Tan, T.Z.; Chiu, D.S.; Kommoss, S.; Leong, H.S.; Chen, S.; Intermaggio, M.P.; et al. Development and Validation of the Gene Expression Predictor of High-Grade Serous Ovarian Carcinoma Molecular SubTYPE (PrOTYPE). Clin. Cancer Res. 2020, 26, 5411–5423. [Google Scholar] [CrossRef] [PubMed]
- Bodelon, C.; Keith Killian, J.; Sampson, J.N.; Anderson, W.F.; Matsuno, R.; Brinton, L.A.; Lissowska, J.; Anglesio, M.S.; Bowtell, D.D.L.; Doherty, J.A.; et al. Molecular Classification of Epithelial Ovarian Cancer Based on Methylation Profiling: Evidence for Survival Heterogeneity. Clin. Cancer Res. 2019, 25, 5937–5946. [Google Scholar] [CrossRef] [PubMed]
- Rusan, M.; Andersen, R.F.; Jakobsen, A.; Steffensen, K.D. Circulating HOXA9-Methylated Tumour DNA: A Novel Biomarker of Response to Poly (ADP-Ribose) Polymerase Inhibition in BRCA-Mutated Epithelial Ovarian Cancer. Eur. J. Cancer 2020, 125, 121–129. [Google Scholar] [CrossRef]
- Heinze, K.; Rengsberger, M.; Gajda, M.; Jansen, L.; Osmers, L.; Oliveira-Ferrer, L.; Schmalfeldt, B.; Dürst, M.; Häfner, N.; Runnebaum, I.B. CAMK2N1/RUNX3 Methylation Is an Independent Prognostic Biomarker for Progression-Free and Overall Survival of Platinum-Sensitive Epithelial Ovarian Cancer Patients. Clin. Epigenetics 2021, 13, 15. [Google Scholar] [CrossRef]
- Calanca, N.; Abildgaard, C.; Rainho, C.A.; Rogatto, S.R. The Interplay between Long Noncoding RNAs and Proteins of the Epigenetic Machinery in Ovarian Cancer. Cancers 2020, 12, 2701. [Google Scholar] [CrossRef]
- Gokulnath, P.; de Cristofaro, T.; Manipur, I.; Di Palma, T.; Soriano, A.A.; Guarracino, M.R.; Zannini, M. Long Non-Coding RNA HAND2-AS1 Acts as a Tumor Suppressor in High-Grade Serous Ovarian Carcinoma. Int. J. Mol. Sci. 2020, 21, 4059. [Google Scholar] [CrossRef]
- Bhardwaj, V.; Tan, Y.Q.; Wu, M.M.; Ma, L.; Zhu, T.; Lobie, P.E.; Pandey, V. Long Non-Coding RNAs in Recurrent Ovarian Cancer: Theranostic Perspectives. Cancer Lett. 2021, 502, 97–107. [Google Scholar] [CrossRef]
- Sun, D.; Fan, X.H. LncRNA SNHG12 Accelerates the Progression of Ovarian Cancer via Absorbing MiRNA-129 to Upregulate SOX4. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 2345–2352. [Google Scholar] [CrossRef]
- Zheng, J.; Guo, J.; Cao, B.; Zhou, Y.; Tong, J. Identification and Validation of LncRNAs Involved in M6A Regulation for Patients with Ovarian Cancer. Cancer Cell Int. 2021, 21, 363. [Google Scholar] [CrossRef]
- Duan, M.; Fang, M.; Wang, C.; Wang, H.; Li, M. LncRNA EMX2OS Induces Proliferation, Invasion and Sphere Formation of Ovarian Cancer Cells via Regulating the MiR-654-3p/AKT3/PD-L1 Axis. Cancer Manag. Res. 2020, 12, 2141–2154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, M.; Ling, W.; Ruan, Z. Long Non-Coding RNA SNHG12 Promotes Immune Escape of Ovarian Cancer Cells through Their Crosstalk with M2 Macrophages. Aging 2020, 12, 17122–17136. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Y.; Wang, X.; Yang, Q. Identification of a Six-Immune-Related Long Non-Coding RNA Signature for Predicting Survival and Immune Infiltrating Status in Breast Cancer. Front. Genet. 2020, 11, 680. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, H.; Wei, Z.; Gou, X.; Liang, S.; Liu, F. A Novel Prognostic Model Based on Autophagy-Related Long Non-Coding RNAs for Clear Cell Renal Cell Carcinoma. Front. Oncol. 2021, 11, 2924. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.C.; Zhang, M.Z.; Shi, M.Y.; Zhang, T.T.; Zhang, Z.N.; Cui, Q.B.; Yang, S.L.; Li, Z.Z. A Survival-Related Competitive Endogenous RNA Network of Prognostic LncRNAs, MiRNAs, and MRNAs in Wilms Tumor. Front. Oncol. 2021, 11, 316. [Google Scholar] [CrossRef]
- Zimta, A.A.; Tigu, A.B.; Braicu, C.; Stefan, C.; Ionescu, C.; Berindan-Neagoe, I. An Emerging Class of Long Non-Coding RNA with Oncogenic Role Arises From the SnoRNA Host Genes. Front. Oncol. 2020, 10, 389. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Sun, W.; Wang, Z.; Dong, W.; He, L.; Zhang, T.; Zhang, H. Long Non-Coding Small Nucleolar RNA Host Genes (SNHGs) in Endocrine-Related Cancers. Onco Targets Ther. 2020, 13, 7699–7717. [Google Scholar] [CrossRef]
- Dong, J.; Wang, Q.; Li, L.; Xiao-jin, Z. Upregulation of Long Non-Coding RNA Small Nucleolar RNA Host Gene 12 Contributes to Cell Growth and Invasion in Cervical Cancer by Acting as a Sponge for MiR-424-5p. Cell Physiol. Biochem. 2018, 45, 2086–2094. [Google Scholar] [CrossRef]
- Lai, S.; Guan, H.; Liu, J.; Huang, L.; Hu, X.; Chen, Y.; Wu, Y.; Wang, Y.; Yang, Q.; Zhou, J. Long Noncoding RNA SNHG12 Modulated by Human Papillomavirus 16 E6/E7 Promotes Cervical Cancer Progression via ERK/Slug Pathway. J. Cell. Physiol. 2020, 235, 7911–7922. [Google Scholar] [CrossRef]
- Ding, S.; Qu, W.; Jiao, Y.; Zhang, J.; Zhang, C.; Dang, S. LncRNA SNHG12 Promotes the Proliferation and Metastasis of Papillary Thyroid Carcinoma Cells through Regulating Wnt/β-Catenin Signaling Pathway. Cancer Biomark. 2018, 22, 217–226. [Google Scholar] [CrossRef]
- Lan, T.; Ma, W.; Hong, Z.; Wu, L.; Chen, X.; Yuan, Y. Long Non-Coding RNA Small Nucleolar RNA Host Gene 12 (SNHG12) Promotes Tumorigenesis and Metastasis by Targeting MiR-199a/b-5p in Hepatocellular Carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; He, C.; Yang, Z.; Li, S.; Qiao, L.; Fang, L. Dysregulation of Long Non-Coding RNA SNHG12 Alters the Viability, Apoptosis, and Autophagy of Prostate Cancer Cells by Regulating MiR-195/CCNE1 Axis. Int. J. Clin. Exp. Pathol. 2019, 12, 1272–1283. [Google Scholar] [PubMed]
- Cheng, G.; Song, Z.; Liu, Y.; Xiao, H.; Ruan, H.; Cao, Q.; Wang, K.; Xiao, W.; Xiong, Z.; Liu, D.; et al. Long Noncoding RNA SNHG12 Indicates the Prognosis of Prostate Cancer and Accelerates Tumorigenesis via Sponging MiR-133b. J. Cell. Physiol. 2020, 235, 1235–1246. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Wang, S.; Liang, X.; Wang, C.; Peng, B. Oncogenic Role of Long Non-coding RNA SNHG12 in Gastric Cancer Cells by Targeting MiR-16. Exp. Ther. Med. 2019, 18, 199–208. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, S.; Yu, Y.; Shi, Y.; Zheng, H. Knockdown of SNHG12 Suppresses Tumor Metastasis and Epithelial-Mesenchymal Transition via the Slug/ZEB2 Signaling Pathway by Targeting MiR-218 in NSCLC. Oncol. Lett. 2019, 17, 2356–2364. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.; Xu, J.; Zuo, Z.; Liu, Y.; Yan, F.; Han, C. Downregulation of LncRNA SNHG12 Reversed IGF1R-Induced Osteosarcoma Metastasis and Proliferation by Targeting MiR-195-5p. Gene 2020, 726, 144145. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, Y.; Liu, H.; Chen, W.; Fan, H.-N.; Zhang, J.; Zhu, J.-S. The Long Non-Coding RNA SNHG12 Promotes Gastric Cancer by Activating the Phosphatidylinositol 3-Kinase/AKT Pathway. Aging 2019, 11, 10902–10922. [Google Scholar] [CrossRef]
- Dong, Y.; Wang, G. Knockdown of LncRNA SNHG12 Suppresses Cell Proliferation, Migration and Invasion in Breast Cancer by Sponging MiR-451a. Int. J. Clin. Exp. Pathol. 2020, 13, 393–402. [Google Scholar]
- Wang, X.; Qi, G.; Zhang, J.; Wu, J.; Zhou, N.; Li, L.; Ma, J. Knockdown of Long Noncoding RNA Small Nucleolar RNA Host Gene 12 Inhibits Cell Growth and Induces Apoptosis by Upregulating MiR-138 in Nonsmall Cell Lung Cancer. DNA Cell Biol. 2017, 36, 892–900. [Google Scholar] [CrossRef]
- Zhou, S.; Yu, L.; Xiong, M.; Dai, G. LncRNA SNHG12 Promotes Tumorigenesis and Metastasis in Osteosarcoma by Upregulating Notch2 by Sponging MiR-195-5p. Biochem. Biophys. Res. Commun. 2018, 495, 1822–1832. [Google Scholar] [CrossRef]
- Lu, C.; Wei, Y.; Wang, X.; Zhang, Z.; Yin, J.; Li, W.; Chen, L.; Lyu, X.; Shi, Z.; Yan, W.; et al. DNA-Methylation-Mediated Activating of LncRNA SNHG12 Promotes Temozolomide Resistance in Glioblastoma. Mol. Cancer 2020, 19, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Cheng, G.; Huang, Z.; Bao, L.; Liu, J.; Wang, C.; Xiong, Z.; Zhou, L.; Xu, T.; Liu, D.; et al. Long Noncoding RNA SNHG12 Promotes Tumour Progression and Sunitinib Resistance by Upregulating CDCA3 in Renal Cell Carcinoma. Cell Death Dis. 2020, 11, 515. [Google Scholar] [CrossRef] [PubMed]
- Lino Cardenas, C.L.; Kessinger, C.W.; Cheng, Y.; MacDonald, C.; MacGillivray, T.; Ghoshhajra, B.; Huleihel, L.; Nuri, S.; Yeri, A.S.; Jaffer, F.A.; et al. An HDAC9-MALAT1-BRG1 Complex Mediates Smooth Muscle Dysfunction in Thoracic Aortic Aneurysm. Nat. Commun. 2018, 9, 1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, X.; Chen, X.; Xue, H.; Tang, Y.; Zhang, P.; Kang, Q.; Hao, Y.; Chen, R.; Zhao, Y.; He, S. NPInter v4.0: An Integrated Database of NcRNA Interactions. Nucleic Acids Res. 2019, 48, D160–D165. [Google Scholar] [CrossRef]
- Brancolini, C.; Di Giorgio, E.; Formisano, L.; Gagliano, T. Quis Custodiet Ipsos Custodes (Who Controls the Controllers)? Two Decades of Studies on HDAC9. Life 2021, 11, 90. [Google Scholar] [CrossRef]
- Khan, M.A.; Vikramdeo, K.S.; Sudan, S.K.; Singh, S.; Wilhite, A.; Dasgupta, S.; Rocconi, R.P.; Singh, A.P. Platinum-Resistant Ovarian Cancer: From Drug Resistance Mechanisms to Liquid Biopsy-Based Biomarkers for Disease Management. Semin. Cancer Biol. 2021, 77, 99–109. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abildgaard, C.; do Canto, L.M.; Rainho, C.A.; Marchi, F.A.; Calanca, N.; Waldstrøm, M.; Steffensen, K.D.; Rogatto, S.R. The Long Non-Coding RNA SNHG12 as a Mediator of Carboplatin Resistance in Ovarian Cancer via Epigenetic Mechanisms. Cancers 2022, 14, 1664. https://doi.org/10.3390/cancers14071664
Abildgaard C, do Canto LM, Rainho CA, Marchi FA, Calanca N, Waldstrøm M, Steffensen KD, Rogatto SR. The Long Non-Coding RNA SNHG12 as a Mediator of Carboplatin Resistance in Ovarian Cancer via Epigenetic Mechanisms. Cancers. 2022; 14(7):1664. https://doi.org/10.3390/cancers14071664
Chicago/Turabian StyleAbildgaard, Cecilie, Luisa Matos do Canto, Cláudia Aparecida Rainho, Fabio Albuquerque Marchi, Naiade Calanca, Marianne Waldstrøm, Karina Dahl Steffensen, and Silvia Regina Rogatto. 2022. "The Long Non-Coding RNA SNHG12 as a Mediator of Carboplatin Resistance in Ovarian Cancer via Epigenetic Mechanisms" Cancers 14, no. 7: 1664. https://doi.org/10.3390/cancers14071664
APA StyleAbildgaard, C., do Canto, L. M., Rainho, C. A., Marchi, F. A., Calanca, N., Waldstrøm, M., Steffensen, K. D., & Rogatto, S. R. (2022). The Long Non-Coding RNA SNHG12 as a Mediator of Carboplatin Resistance in Ovarian Cancer via Epigenetic Mechanisms. Cancers, 14(7), 1664. https://doi.org/10.3390/cancers14071664