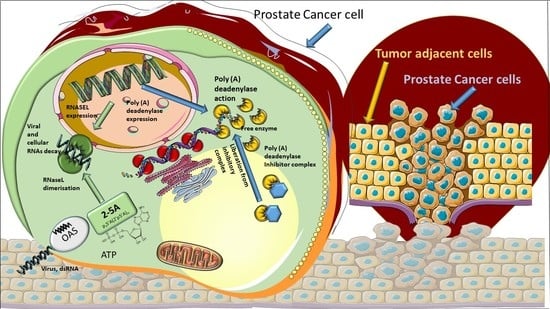
Template-Independent Poly(A)-Tail Decay and RNASEL as Potential Cellular Biomarkers for Prostate Cancer Development
Abstract
:Simple Summary
Abstract
1. Introduction
2. Patients and Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrano, N.A.; Anscher, M.S. Favorable vs. Unfavorable Intermediate-Risk Prostate Cancer: A Review of the New Classification System and Its Impact on Treatment Recommendations. Oncology 2016, 30, 229–236. [Google Scholar] [PubMed]
- Kretschmer, A.; Tilki, D. Biomarkers in prostate cancer—Current clinical utility and future perspectives. Crit. Rev. Oncol. Hematol. 2017, 120, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Cox, P.M.; Goding, C.R. Transcription and cancer. Br. J. Cancer 1991, 63, 651–662. [Google Scholar] [CrossRef] [Green Version]
- Garneau, N.L.; Wilusz, J.; Wilusz, C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol. 2007, 8, 113–126. [Google Scholar] [CrossRef]
- Parker, R.; Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004, 11, 121–127. [Google Scholar] [CrossRef]
- Chen, C.Y.; Shyu, A.B. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip. Rev. RNA 2011, 2, 167–183. [Google Scholar] [CrossRef] [Green Version]
- Goldstrohm, A.C.; Wickens, M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008, 9, 337–344. [Google Scholar] [CrossRef]
- Arafat, M.; Sperling, R.A. Quality Control Mechanism of Splice Site Selection Abrogated under Stress and in Cancer. Cancers 2022, 14, 1750. [Google Scholar] [CrossRef]
- Lejeune, F. Nonsense-mediated mRNA decay at the crossroads of many cellular pathways. BMB Rep. 2017, 50, 175–185. [Google Scholar] [CrossRef] [Green Version]
- Sandberg, R.; Neilson, J.R.; Sarma, A.; Sharp, P.A.; Burge, C.B. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 2008, 320, 1643–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, A.R.; Bos, A.; Diosdado, B.; Rooijers, K.; Elkon, R.; Bolijn, A.S.; Carvalho, B.; Meijer, G.A.; Agami, R. Alternative cleavage and polyadenylation during colorectal cancer development. Clin. Cancer Res. 2012, 18, 5256–5266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayr, C.; Bartel, D.P. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009, 138, 673–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, R.; Shi, Y. The polyadenylation code: A unified model for the regulation of mRNA alternative polyadenylation. J. Zhejiang Univ. Sci. B 2014, 15, 429–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sciarra, A.; Gentilucci, A.; Salciccia, S.; Pierella, F.; Del Bianco, F.; Gentile, V.; Silvestri, I.; Cattarino, S. Prognostic value of inflammation in prostate cancer progression and response to therapeutic: A critical review. J. Inflamm. 2016, 13, 35. [Google Scholar] [CrossRef] [Green Version]
- Gandaglia, G.; Briganti, A.; Gontero, P.; Mondaini, N.; Novara, G.; Salonia, A.; Sciarra, A.; Montorsi, F. The role of chronic prostatic inflammation in the pathogenesis and progression of benign prostatic hyperplasia. BJU Int. 2013, 112, 432–441. [Google Scholar] [CrossRef]
- Wang, S.; Ji, Y.; Chen, Y.; Du, P.; Cao, Y.; Yang, X.; Ma, J.; Yu, Z.; Yang, Y. The Values of Systemic Immune-Inflammation Index and Neutrophil-Lymphocyte Ratio in the Localized Prostate Cancer and Benign Prostate Hyperplasia: A Retrospective Clinical Study. Front. Oncol. 2022, 11, 812319. [Google Scholar] [CrossRef]
- Mughees, M.; Kaushal, J.B.; Sharma, G.; Wajid, S.; Batra, S.K.; Siddiqui, J.A. Chemokines and Cytokines: Axis and Allies in Prostate Cancer Pathogenesis. Semin. Cancer Biol. 2022. [Google Scholar] [CrossRef]
- Silverman, R.H. Implications for RNase L in prostate cancer biology. Biochemistry 2003, 42, 1805–1812. [Google Scholar] [CrossRef]
- Silverman, R.H. A scientific journey through the 2-5A/RNase L system. Cytokine Growth Factor Rev. 2007, 18, 381–388. [Google Scholar] [CrossRef] [Green Version]
- Casey, G.; Neville, P.J.; Plummer, S.J.; Xiang, Y.; Krumroy, L.M.; Klein, E.A.; Catalona, W.J.; Nupponen, N.; Carpten, J.D.; Trent, J.M.; et al. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat. Genet. 2002, 32, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Rokman, A.; Ikonen, T.; Seppala, E.H.; Nupponen, N.; Autio, V.; Mononen, N.; Bailey-Wilson, J.; Trent, J.; Carpten, J.; Matikainen, M.P.; et al. Germline alterations of the RNASEL gene, a candidate HPC1 gene at 1q25, in patients and families with prostate cancer. Am. J. Hum. Genet. 2002, 70, 1299–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiklund, F.; Jonsson, B.A.; Brookes, A.J.; Stromqvist, L.; Adolfsson, J.; Emanuelsson, M.; Adami, H.O.; Augustsson-Balter, K.; Gronberg, H. Genetic analysis of the RNASEL gene in hereditary, familial, and sporadic prostate cancer. Clin. Cancer Res. 2004, 10, 7150–7156. [Google Scholar] [CrossRef] [Green Version]
- Maier, C.; Haeusler, J.; Herkommer, K.; Vesovic, Z.; Hoegel, J.; Vogel, W.; Paiss, T. Mutation screening and association study of RNASEL as a prostate cancer susceptibility gene. Br. J. Cancer 2005, 92, 1159–1164. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Sun, R. Evidence from 40 Studies that 2 Common Single-Nucleotide Polymorphisms (SNPs) of RNASEL Gene Affect Prostate Cancer Susceptibility: A Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)-Compliant Meta-Analysis. Med. Sci. Monit. 2019, 25, 8315–8325. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Wang, Z.; Murakami, J.; Plummer, S.; Klein, E.A.; Carpten, J.D.; Trent, J.M.; Isaacs, W.B.; Casey, G.; Silverman, R.H. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′,5′-oligoadenylates. Cancer Res. 2003, 63, 6795–6801. [Google Scholar] [PubMed]
- Kocic, G.; Bjelakovic, G.; Pavlovic, D.; Jevtovic, T.; Pavlovic, V.; Sokolovic, D.; Basic, J.; Cekic, S.; Cvetkovic, T.; Kocic, R.; et al. Protective effect of interferon-alpha on the DNA- and RNA-degrading pathway in anti-Fas-antibody induced apoptosis. Hepatol. Res. 2007, 37, 637–646. [Google Scholar] [CrossRef]
- Kocic, G.; Veljkovic, A.; Kocic, H.; Colic, M.; Mihajlovic, D.; Tomovic, K.; Stojanovic, S.; Smelcerovic, A. Depurinized milk downregulates rat thymus MyD88/Akt/p38 function, NF-κB-mediated inflammation, caspase-1 activity but not the endonuclease pathway:in vitro/in vivo study. Sci. Rep. 2017, 7, 41971. [Google Scholar] [CrossRef] [Green Version]
- Kocic, G.; Pavlovic, R.; Nikolic, G.; Veljkovic, A.; Panseri, S.; Chiesa, L.M.; Andjelkovic, T.; Jevtovic-Stoimenov, T.; Sokolovic, D.; Cvetkovic, T.; et al. Effect of commercial or depurinized milk on rat liver growth-regulatory kinases, nuclear factor-kappa B, and endonuclease in experimental hyperuricemia: Comparison with allopurinol therapy. J. Dairy Sci. 2014, 97, 4029–4042. [Google Scholar] [CrossRef] [Green Version]
- Dickson, K.A.; Haigis, M.C.; Raines, R.T. Ribonuclease inhibitor: Structure and function. Prog. Nucleic Acid Res. Mol. Biol. 2005, 80, 349–374. [Google Scholar]
- Cho, S.W.; Joshi, J.G. Ribonuclease inhibitor from pig brain: Purification, characterization, and direct spectrophotometric assay. Anal. Biochem. 1989, 176, 175–179. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosenbrough, N.J.; Farr, A.J.; Randall, R.J. Protein measurement with the pholin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Hartwell, L.; Mankoff, D.; Paulovich, A.; Ramsey, S.; Swisher, E.M. Cancer biomarkers: A systems approach. Nat. Biotechnol. 2006, 24, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Madu, C.O.; Lu, Y. Novel diagnostic biomarkers for prostate cancer. J. Cancer 2010, 1, 150–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.N.; Yan, Y.B. Depletion of poly(A)-specific ribonuclease (PARN) inhibits proliferation of human gastric cancer cells by blocking cell cycle progression. Biochim. Biophys. Acta 2015, 1853, 522–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maragozidis, P.; Karangeli, M.; Labrou, M.; Dimoulou, G.; Papaspyrou, K.; Salataj, E.; Pournaras, S.; Matsouka, P.; Gourgoulianis, K.I.; Balatsos, N. Alterations of deadenylase expression in acute leukemias: Evidence for poly(a)-specific ribonuclease as a potential biomarker. Acta Haematol. 2012, 128, 39–46. [Google Scholar] [CrossRef]
- Pavlopoulou, A.; Vlachakis, D.; Balatsos, N.A.; Kossida, S. A comprehensive phylogenetic analysis of deadenylases. Evol. Bioinform. Online 2013, 9, 491–497. [Google Scholar] [CrossRef] [Green Version]
- Virtanen, A.; Henriksson, N.; Nilsson, P.; Nissbeck, M. Poly(A)-specific ribonuclease (PARN): An allosterically regulated, processive and mRNA cap-interacting deadenylase. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 192–209. [Google Scholar] [CrossRef]
- Godwin, A.R.; Kojima, S.; Green, C.B.; Wilusz, J. Kiss your tail goodbye: The role of PARN, Nocturnin, and Angel deadenylases in mRNA biology. Biochim. Biophys. Acta 2013, 1829, 571–579. [Google Scholar] [CrossRef] [Green Version]
- Balatsos, N.A.; Maragozidis, P.; Anastasakis, D.; Stathopoulos, C. Modulation of poly(A)-specific ribonuclease (PARN): Current knowledge and perspectives. Curr. Med. Chem. 2012, 19, 4838–4849. [Google Scholar] [CrossRef]
- Balatsos, N.A.; Nilsson, P.; Mazza, C.; Cusack, S.; Virtanen, A. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC). J. Biol. Chem. 2006, 281, 4517–4522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maragozidis, P.; Papanastasi, E.; Scutelnic, D.; Totomi, A.; Kokkori, I.; Zarogiannis, S.G.; Kerenidi, T.; Gourgoulianis, K.I.; Balatsos, N.A.A. Poly(A)-specific ribonuclease and Nocturnin in squamous cell lung cancer: Prognostic value and impact on gene expression. Mol. Cancer 2015, 14, 187. [Google Scholar] [CrossRef] [PubMed]
- Pandolfi, P.P. Aberrant mRNA translation in cancer pathogenesis: An old concept revisited comes finally of age. Oncogene 2004, 23, 3134–3137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, J.Z.; Hong, A.; Lilly, M.A.; Lehmann, R. Twin, a CCR4 homolog, regulates cyclin poly(A) tail length to permit Drosophila oogenesis. Development 2005, 132, 1165–1174. [Google Scholar] [CrossRef] [Green Version]
- Berthet, C.; Morera, A.-M.; Asensio, M.-J.; Chauvin, M.-A.; Morel, A.-P.; Dijoud, F.; Magaud, J.-P.; Durand, P.; Rouault, J.-P. CCR4-associated factor CAF1 is an essential factor for spermatogenesis. Mol. Cell. Biol. 2004, 24, 5808–5820. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Yao, R.; Ogawa, T.; Suzuki, T.; Ito, C.; Tsunekawa, N.; Inoue, K.; Ajima, R.; Miyasaka, T.; Yoshida, Y.; et al. Oligo-astheno-teratozoospermia in mice lacking CNOT7, a regulator of retinoid X receptor β. Nat. Genet. 2004, 36, 528–533. [Google Scholar] [CrossRef]
- Fominaya, J.M.; Hofsteenge, J. Inactivation of ribonuclease inhibitor by thiol–disulfide exchange. J. Biol. Chem. 1992, 267, 24655–24660. [Google Scholar] [CrossRef]
- Hofsteenge, J.; Kieffer, B.; Matthies, R.; Hemmings, B.A.; Stone, S.R. Amino acid sequence of the ribonuclease inhibitor from porcine liver reveals the presence of leucine-rich repeats. Biochemistry 1988, 27, 8537–8544. [Google Scholar] [CrossRef]
- Nadano, D.; Yasuda, T.; Takeshita, H.; Uchide, K.; Kishi, K. Purification and characterization of human brain ribonuclease inhibitor. Arch. Biochem. Biophys. 1994, 312, 421–428. [Google Scholar] [CrossRef]
- Futami, J.; Tsushima, Y.; Murato, Y.; Tada, H.; Sasaki, J.; Seno, M.; Yamada, H. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA Cell Biol. 1997, 16, 413–419. [Google Scholar] [CrossRef]
- Burton, L.E.; Fucci, N.P. Ribonuclease inhibitors from the liver of five mammalian species. Int. J. Pept. Protein Res. 1982, 19, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Ferreras, M.; Gavilanes, J.G.; Lopez-Otin, C.; Garcia-Segura, J.M. Thiol–disulfide exchange of ribonuclease inhibitor bound to ribonuclease A. Evidence of active inhibitor-bound ribonuclease. J. Biol. Chem. 1995, 270, 28570–28578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjelakovic, G.; Pavlovic, D.; Nikolic, J.; Kocic, G.; Stankovic, B.; Bjelakovic, B. Effect of pyridoxine on the alkaline ribonuclease activity in the liver of dexamethasone treated rats. Facta Univ. Ser. Med. Biol. 1997, 4, 17–20. [Google Scholar]
- Kraft, N.; Shortman, K. The phylogeny of the ribonuclease-ribonuclease inhibitor system: Its distribution in tissues and its response during leukaemogenesis and aging. Aust. J. Biol. Sci. 1970, 23, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Veljković, A.; Hadži-Dokić, J.; Sokolović, D.; Bašić, D.; Veličković-Janković, L.; Stojanović, M.; Popović, D.; Kocić, G. Xanthine Oxidase/Dehydrogenase Activity as a Source of Oxidative Stress in Prostate Cancer Tissue. Diagnostics 2020, 10, 668. [Google Scholar] [CrossRef] [PubMed]
- Karan, D.; Dubey, S. From Inflammation to Prostate Cancer: The Role of inflammasomes. Adv. Urol. 2016, 2016, 3140372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ammirante, M.; Luo, J.L.; Grivennikov, S.; Nedospasov, S.; Karin, M. B-cell-derived lymphotoxin promotes castration-resistant prostate cancer. Nature 2010, 464, 302–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurel, B.; Lucia, M.S.; Thompson, I.M.; Goodman, P.J., Jr.; Tangen, C.M.; Kristal, A.R.; Parnes, H.L.; Hoque, A.; Lippman, S.M.; Sutcliffe, S.; et al. Chronic inflammation in benign prostate tissue is associated with high-grade prostate cancer in the placebo arm of the prostate cancer prevention trial. Cancer Epidemiol. Biomark. Prev. 2014, 23, 847–856. [Google Scholar] [CrossRef] [Green Version]
- Carpten, J.; Nupponen, N.; Isaacs, S.; Sood, R.; Robbins, C.; Xu, J.; Faruque, M.; Moses, T.; Ewing, C.; Gillanders, E.; et al. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat. Genet. 2002, 30, 181–184. [Google Scholar] [CrossRef]








| Investigated Parameters | n% |
|---|---|
| Age | |
| <70 | 28 (60.87%) |
| >70 | 18 (39.13%) |
| Tumor stage | |
| II | 34 (73.91%) |
| III | 12 (26.09%) |
| pN—lymph node metastasis | |
| NO | 20 (43.48%) |
| NX | 26 (56.52%) |
| pM-distant metastasis | |
| MO | 22 (47.83%) |
| MX | 24 (52.17%) |
| Gleason score | |
| 3 + 3 | 15 (32.6%) |
| 3 + 4 | 18 (39.13%) |
| 4 + 3 | 9 (19.56%) |
| 3 + 5 | 2 (4.35%) |
| 4 + 4 | 2 (4.35%) |
| PSA (ng/mL) | |
| <10 | 28 (60.87%) |
| >10 | 18 (39.13%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kocić, G.; Hadzi-Djokić, J.; Veljković, A.; Roumeliotis, S.; Janković-Veličković, L.; Šmelcerović, A. Template-Independent Poly(A)-Tail Decay and RNASEL as Potential Cellular Biomarkers for Prostate Cancer Development. Cancers 2022, 14, 2239. https://doi.org/10.3390/cancers14092239
Kocić G, Hadzi-Djokić J, Veljković A, Roumeliotis S, Janković-Veličković L, Šmelcerović A. Template-Independent Poly(A)-Tail Decay and RNASEL as Potential Cellular Biomarkers for Prostate Cancer Development. Cancers. 2022; 14(9):2239. https://doi.org/10.3390/cancers14092239
Chicago/Turabian StyleKocić, Gordana, Jovan Hadzi-Djokić, Andrej Veljković, Stefanos Roumeliotis, Ljubinka Janković-Veličković, and Andrija Šmelcerović. 2022. "Template-Independent Poly(A)-Tail Decay and RNASEL as Potential Cellular Biomarkers for Prostate Cancer Development" Cancers 14, no. 9: 2239. https://doi.org/10.3390/cancers14092239
APA StyleKocić, G., Hadzi-Djokić, J., Veljković, A., Roumeliotis, S., Janković-Veličković, L., & Šmelcerović, A. (2022). Template-Independent Poly(A)-Tail Decay and RNASEL as Potential Cellular Biomarkers for Prostate Cancer Development. Cancers, 14(9), 2239. https://doi.org/10.3390/cancers14092239