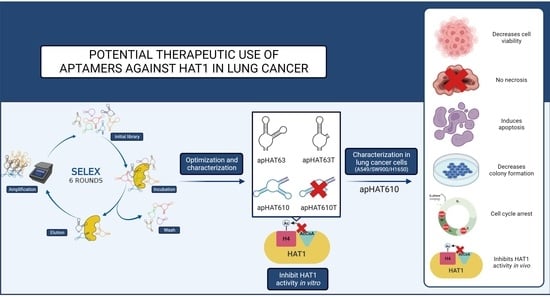
Potential Therapeutic Use of Aptamers against HAT1 in Lung Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Expression and Purification of Recombinant HAT1
2.3. In Vitro Selection
2.4. Aptamer Cloning, Sequencing, and Secondary Structure Prediction
2.5. Next-Generation Sequencing
2.6. Enzyme-Linked Oligonucleotide Assay (ELONA)
2.7. Evolution Analysis of the Aptamer Population by qPCR
2.8. In Vitro HAT1 Activity Assay
2.9. Aptamer Stability Assays
2.10. Cell Culture and Protein and RNA Extraction
2.11. Western Blot and Apta-Western Assays
2.12. Cell Viability (MTT) and Cell Death (LDH) Assays
2.13. Flow Cytometric Analysis of Cell Cycle
2.14. Colony-Forming Assays
2.15. Statistical Analysis
3. Results
3.1. Selection and Characterization of High-Affinity Aptamers against HAT1
3.2. Obtaining Unique Sequences That Specifically Recognize HAT1
3.3. Structural Characterization and Optimization of the Selected Aptamers
3.4. The Aptamers Selected against HAT1 Recognize Their Target with High Affinity and Specificity
3.5. Aptamers against HAT1 Inhibit Acetyltransferase Activity In Vitro
3.6. The Aptamers against HAT1 Are Stable in Human Plasma
3.7. Aptamers against HAT1 Inhibit Cell Viability in Lung Tumor Cells
3.8. apHAT610 Triggers Cell Cycle Arrest in Lung Tumor Cells
3.9. apHAT610 Induces Apoptosis in Lung Tumor Cells
3.10. apHAT610 Inhibits the Acetyltransferase Activity of HAT1 in Lung Tumor Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karimpour, M.; Ravanbakhsh, R.; Maydanchi, M.; Rajabi, A.; Azizi, F.; Saber, A. Cancer driver gene and non-coding RNA alterations as biomarkers of brain metastasis in lung cancer: A review of the literature. Biomed. Pharmacother. 2021, 143, 112190. [Google Scholar] [CrossRef] [PubMed]
- Nasim, F.; Sabath, B.F.; Eapen, G.A. Lung Cancer. Med. Clin. N. Am. 2019, 103, 463–473. [Google Scholar] [CrossRef]
- Brownell, J.E.; Allis, C.D. Special HATs for special occasions: Linking histone acetylation to chromatin assembly and gene activation. Curr. Opin. Genet. Dev. 1996, 6, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Parthun, M.R. Hat1: The emerging cellular roles of a type B histone acetyltransferase. Oncogene 2007, 26, 5319–5328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleff, S.; Andrulis, E.D.; Anderson, C.W.; Sternglanz, R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J. Biol. Chem. 1995, 270, 24674–24677. [Google Scholar] [CrossRef] [Green Version]
- Dutnall, R.N.; Tafrov, S.T.; Sternglanz, R.; Ramakrishnan, V. Structure of the histone acetyltransferase Hat1: A paradigm for the GCN5-related N-acetyltransferase superfamily. Cell 1998, 94, 427–438. [Google Scholar] [CrossRef] [Green Version]
- Verreault, A.; Kaufman, P.D.; Kobayashi, R.; Stillman, B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 1998, 8, 96–108. [Google Scholar] [CrossRef] [Green Version]
- Poziello, A.; Nebbioso, A.; Stunnenberg, H.G.; Martens, J.H.; Carafa, V.; Altucci, L. Recent insights into Histone Acetyltransferase-1: Biological function and involvement in pathogenesis. Epigenetics 2020, 16, 838–850. [Google Scholar] [CrossRef]
- Ruiz-García, A.B.; Sendra, R.; Galiana, M.; Pamblanco, M.; Pérez-Ortín, J.E.; Tordera, V. HAT1 and HAT2 proteins are components of a yeast nuclear histone acetyltransferase enzyme specific for free histone H4. J. Biol. Chem. 1998, 273, 12599–12605. [Google Scholar] [CrossRef]
- Sterner, D.E.; Berger, S.L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000, 64, 435–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagamori, I.; Cruickshank, V.A.; Sassone-Corsi, P. Regulation of an RNA granule during spermatogenesis: Acetylation of MVH in the chromatoid body of germ cells. J. Cell. Sci. 2011, 124 Pt 24, 4346–4355. [Google Scholar] [CrossRef] [Green Version]
- Sadler, A.J.; Suliman, B.A.; Yu, L.; Yuan, X.; Wang, D.; Irving, A.T.; Sarvestani, S.T.; Banerjee, A.; Mansell, A.S.; Liu, J.P.; et al. The acetyltransferase HAT1 moderates the NF-kappaB response by regulating the transcription factor PLZF. Nat. Commun. 2015, 6, 6795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imhof, A.; Wolffe, A.P. Purification and properties of the Xenopus Hat1 acetyltransferase: Association with the 14-3-3 proteins in the oocyte nucleus. Biochemistry 1999, 38, 13085–13093. [Google Scholar] [CrossRef]
- Barman, H.K.; Takami, Y.; Ono, T.; Nishijima, H.; Sanematsu, F.; Shibahara, K.-I.; Nakayama, T. Histone acetyltransferase 1 is dispensable for replication-coupled chromatin assembly but contributes to recover DNA damages created following replication blockage in vertebrate cells. Biochem. Biophys. Res. Commun. 2006, 345, 1547–1557. [Google Scholar] [CrossRef]
- Agudelo Garcia, P.A.; Nagarajan, P.; Parthun, M.R. Hat1-Dependent Lysine Acetylation Targets Diverse Cellular Functions. J. Proteome Res. 2020, 19, 1663–1673. [Google Scholar] [CrossRef] [PubMed]
- Poveda, A.; Sendra, R. Site specificity of yeast histone acetyltransferase B complex in vivo. FEBS J. 2008, 275, 2122–2136. [Google Scholar] [CrossRef]
- De Oliveira, M.; De Sibio, M.T.; Mathias, L.S.; Rodrigues, B.M.; Sakalem, M.E.; Nogueira, C.R. Irisin modulates genes associated with severe coronavirus disease (COVID-19) outcome in human subcutaneous adipocytes cell culture. Mol. Cell. Endocrinol. 2020, 515, 110917. [Google Scholar] [CrossRef]
- Demyanenko, S.; Uzdensky, A. Epigenetic Alterations Induced by Photothrombotic Stroke in the Rat Cerebral Cortex: Deacetylation of Histone h3, Upregulation of Histone Deacetylases and Histone Acetyltransferases. Int. J. Mol. Sci. 2019, 20, 2882. [Google Scholar] [CrossRef] [Green Version]
- Demyanenko, S.V.; Dzreyan, V.A.; Uzdensky, A.B. The Expression and Localization of Histone Acetyltransferases HAT1 and PCAF in Neurons and Astrocytes of the Photothrombotic Stroke-Induced Penumbra in the Rat Brain Cortex. Mol. Neurobiol. 2020, 57, 3219–3227. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, M.; Xie, W.; Lan, G.; Cheng, H.-P.; Gong, D.; Huang, C.; Lv, Y.-C.; Yao, F.; Tan, Y.-L.; et al. MiR-486 regulates cholesterol efflux by targeting HAT1. Biochem. Biophys. Res. Commun. 2016, 472, 418–424. [Google Scholar] [CrossRef]
- Pinto, B.G.G.; Oliveira, A.E.R.; Singh, Y.; Jimenez, L.; A Gonçalves, A.N.; Ogava, R.L.T.; Creighton, R.; Peron, J.P.S.; Nakaya, H.I. ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19. J. Infect. Dis. 2020, 222, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Sundar, I.K.; Rahman, I. Gene expression profiling of epigenetic chromatin modification enzymes and histone marks by cigarette smoke: Implications for COPD and lung cancer. Am. J. Physiol. Lung. Cell. Mol. Physiol. 2016, 311, L1245–L1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlad, M.-L.; Manea, S.-A.; Lazar, A.-G.; Raicu, M.; Muresian, H.; Simionescu, M.; Manea, A. Histone Acetyltransferase-Dependent Pathways Mediate Upregulation of NADPH Oxidase 5 in Human Macrophages under Inflammatory Conditions: A Potential Mechanism of Reactive Oxygen Species Overproduction in Atherosclerosis. Oxid. Med. Cell. Longev. 2019, 2019, 3201062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Liu, K.; Fang, B.A.M.; Wu, H.; Li, F.; Xiang, X.; Tang, W.; Zhao, G.; Lin, L.; Bao, S.; et al. Identification of acetyltransferase genes (HAT1 and KAT8) regulating HBV replication by RNAi screening. Cell Biosci. 2015, 5, 66. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Feng, J.; Liu, Y.; Zhao, M.; Yuan, Y.; Yuan, H.; Yun, H.; Sun, M.; Bu, Y.; Liu, L.; et al. HAT1 signaling confers to assembly and epigenetic regulation of HBV cccDNA minichromosome. Theranostics 2019, 9, 7345–7358. [Google Scholar] [CrossRef]
- Yuan, Y.; Miao, Y.; Qian, L.; Zhang, Y.; Liu, C.; Liu, J.; Zuo, Y.; Feng, Q.; Guo, T.; Zhang, L.; et al. Targeting UBE4A Revives Viperin Protein in Epithelium to Enhance Host Antiviral Defense. Mol. Cell 2020, 77, 734–747.e7. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, Z.; Kong, L.; Gao, H.; Zhang, Y.; Zheng, Y.; Wan, Y. miRNA-486-5p Promotes COPD Progression by Targeting HAT1 to Regulate the TLR4-Triggered Inflammatory Response of Alveolar Macrophages. Int. J. Chronic Obstr. Pulm. Dis. 2020, 15, 2991–3001. [Google Scholar] [CrossRef]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A.J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 2017, 8, 1077. [Google Scholar] [CrossRef] [Green Version]
- Fan, P.; Zhao, J.; Meng, Z.; Wu, H.; Wang, B.; Wu, H.; Jin, X. Overexpressed histone acetyltransferase 1 regulates cancer immunity by increasing programmed death-ligand 1 expression in pancreatic cancer. J. Exp. Clin. Cancer Res. 2019, 38, 47. [Google Scholar] [CrossRef]
- Hong, Z.; Xiang, Z.; Zhang, P.; Wu, Q.; Xu, C.; Wang, X.; Shi, G.; Hong, Z.; Wu, D. Histone acetyltransferase 1 upregulates androgen receptor expression to modulate CRPC cell resistance to enzalutamide. Clin. Transl. Med. 2021, 11, e495. [Google Scholar] [CrossRef] [PubMed]
- Min, S.K.; Koh, Y.H.; Park, Y.; Kim, H.J.; Seo, J.; Park, H.R.; Cho, S.J.; Kim, I.S. Expression of HAT1 and HDAC1, 2, 3 in Diffuse Large B-Cell Lymphomas, Peripheral T-Cell Lymphomas, and NK/T-Cell Lymphomas. Korean J. Pathol. 2012, 46, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.; Hou, J.; Wang, Q.; Yao, L.; Xu, S.; Ge, D. RNAi screening identifies HAT1 as a potential drug target in esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 3898–3907. [Google Scholar]
- Chrun, E.D.S.; Modolo, F.; Vieira, D.S.C.; Borges-Júnior, Á.L.S.; Castro, R.G.; Daniel, F.I. Immunoexpression of HDAC1, HDAC2, and HAT1 in actinic cheilitis and lip squamous cell carcinoma. Oral Dis. 2017, 23, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Miao, B.-P.; Zhang, R.-S.; Yang, G.; Sun, J.-J.; Tang, Y.-Y.; Liang, W.-F.; Liu, T.; Wen, Z.; Yang, P.-C.; Nie, G.-H. Histone acetyltransferase 1 up regulates Bcl2L12 expression in nasopharyngeal cancer cells. Arch. Biochem. Biophys. 2018, 646, 72–79. [Google Scholar] [CrossRef]
- Xia, P.; Gu, R.; Zhang, W.; Shao, L.; Li, F.; Wu, C.; Sun, Y. MicroRNA-377 exerts a potent suppressive role in osteosarcoma through the involvement of the histone acetyltransferase 1-mediated Wnt axis. J. Cell. Physiol. 2019, 234, 22787–22798. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Chao, A.; Wu, R.-C.; Lee, L.-Y.; Ueng, S.-H.; Tsai, C.-L.; Lee, Y.-S.; Peng, M.-T.; Yang, L.-Y.; Huang, H.-J.; et al. Synergistic effects of pazopanib and hyperthermia against uterine leiomyosarcoma growth mediated by downregulation of histone acetyltransferase 1. J. Mol. Med. 2020, 98, 1175–1188. [Google Scholar] [CrossRef]
- Wang, W.; Li, X.; Xu, Y.; Guo, W.; Yu, H.; Zhang, L.; Wang, Y.; Chen, X. Acetylation-stabilized chloride intracellular channel 1 exerts a tumor-promoting effect on cervical cancer cells by activating NF-kappaB. Cell. Oncol. 2021, 44, 557–568. [Google Scholar] [CrossRef]
- JJin, X.; Tian, S.; Li, P. Histone Acetyltransferase 1 Promotes Cell Proliferation and Induces Cisplatin Resistance in Hepatocellular Carcinoma. Oncol. Res. 2017, 25, 939–946. [Google Scholar] [CrossRef]
- Pogribny, I.P.; Tryndyak, V.P.; Muskhelishvili, L.; Rusyn, I.; Ross, S.A. Methyl deficiency, alterations in global histone modifications, and carcinogenesis. J. Nutr. 2007, 137 (Suppl. S1), 216S–222S. [Google Scholar] [CrossRef] [Green Version]
- Seiden-Long, I.M.; Brown, K.R.; Shih, W.; A Wigle, D.; Radulovich, N.; Jurisica, I.; Tsao, M.-S. Transcriptional targets of hepatocyte growth factor signaling and Ki-ras oncogene activation in colorectal cancer. Oncogene 2006, 25, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Gruber, J.J.; Geller, B.; Lipchik, A.M.; Chen, J.; Salahudeen, A.A.; Ram, A.N.; Ford, J.M.; Kuo, C.J.; Snyder, M.P. HAT1 Coordinates Histone Production and Acetylation via H4 Promoter Binding. Mol. Cell 2019, 75, 711–724.e5. [Google Scholar] [CrossRef] [PubMed]
- Gruber, J.J.; Rangarajan, A.; Chou, T.; Geller, B.S.; Banuelos, S.; Greenhouse, R.; Snyder, M.P.; Lipchik, A.M. An acetyl-click screening platform identifies a small molecule inhibitor of Histone Acetyltransferase 1 (HAT1) with anti-tumor activity. BioRxiv 2021. [Google Scholar] [CrossRef]
- Espíndola, M.S.; Soares, L.S.; Galvão-Lima, L.J.; Zambuzi, F.A.; Cacemiro, M.C.; Brauer, V.S.; Marzocchi-Machado, C.M.; Gomes, M.D.S.; Amaral, L.R.; Martins-Filho, O.A.; et al. Epigenetic alterations are associated with monocyte immune dysfunctions in HIV-1 infection. Sci. Rep. 2018, 8, 5505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tafrova, J.I.; Tafrov, S.T. Human histone acetyltransferase 1 (Hat1) acetylates lysine 5 of histone H2A in vivo. Mol. Cell. Biochem. 2014, 392, 259–272. [Google Scholar] [CrossRef]
- Ngo, L.; Brown, T.; Zheng, Y.G. Bisubstrate inhibitors to target histone acetyltransferase 1. Chem. Biol. Drug Des. 2019, 93, 865–873. [Google Scholar] [CrossRef]
- Wan, Q.; Liu, X.; Zu, Y. Oligonucleotide aptamers for pathogen detection and infectious disease control. Theranostics 2021, 11, 9133–9161. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- González, V.M.; Martín, M.E.; Fernández, G.; García-Sacristán, A. Use of Aptamers as Diagnostics Tools and Antiviral Agents for Human Viruses. Pharmaceuticals 2016, 9, 78. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Li, H.; Zhao, L.; Zhang, Y.; Liu, Z. Oligonucleotide aptamers: Recent advances in their screening, molecular conformation and therapeutic applications. Biomed. Pharmacother. 2021, 143, 112232. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug. Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Zhao, J.; Guo, Z.; Liu, Y.; Chen, H.; Chen, Z.; He, N. Applications of Aptamer-Bound Nanomaterials in Cancer Therapy. Biosensors 2021, 11, 344. [Google Scholar] [CrossRef] [PubMed]
- Mayer, G. The chemical biology of aptamers. Angew. Chem. Int. Ed. Engl. 2009, 48, 2672–2689. [Google Scholar] [CrossRef] [PubMed]
- Ciancio, D.R.; Vargas, M.R.; Thiel, W.H.; Bruno, M.A.; Giangrande, P.H.; Mestre, M.B. Aptamers as Diagnostic Tools in Cancer. Pharmaceuticals 2018, 11, 86. [Google Scholar] [CrossRef] [Green Version]
- García-Recio, E.M.; Pinto-Díez, C.; Pérez-Morgado, M.I.; García-Hernández, M.; Fernández, G.; Martín, M.E.; González, V.M. Characterization of MNK1b DNA Aptamers That Inhibit Proliferation in MDA-MB231 Breast Cancer Cells. Mol. Ther. Nucleic Acids 2016, 5, e275. [Google Scholar] [CrossRef] [Green Version]
- Guerra-Pérez, N.; Ramos, E.; García-Hernández, M.; Pinto, C.; Soto, M.; Martín, M.E.; González, V.M. Molecular and Functional Characterization of ssDNA Aptamers that Specifically Bind Leishmania infantum PABP. PLoS ONE 2015, 10, e0140048. [Google Scholar] [CrossRef] [Green Version]
- Pereira, R.L.; Nascimento, I.C.; Santos, A.P.; Ogusuku, I.E.Y.; Lameu, C.; Mayer, G.; Ulrich, H. Aptamers: Novelty tools for cancer biology. Oncotarget 2018, 9, 26934–26953. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Rossi, J.J. Cell-type-specific, Aptamer-functionalized Agents for Targeted Disease Therapy. Mol. Ther. Nucleic Acids 2014, 3, e169. [Google Scholar] [CrossRef]
- Iborra, S.; Soto, M.; Carrión, J.; Alonso, C.; Requena, J.M. Vaccination with a plasmid DNA cocktail encoding the nucleosomal histones of Leishmania confers protection against murine cutaneous leishmaniosis. Vaccine 2004, 22, 3865–3876. [Google Scholar] [CrossRef]
- Morris, K.N.; Jensen, K.B.; Julin, C.M.; Weil, M.; Gold, L. High affinity ligands from in vitro selection: Complex targets. Proc. Natl. Acad. Sci. USA 1998, 95, 2902–2907. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; He, L.; Wang, J.; Fang, X.; Zhang, L. Developing a combined strategy for monitoring the progress of aptamer selection. Analyst 2017, 142, 3136–3139. [Google Scholar] [CrossRef] [PubMed]
- Long, B.H.; Musial, S.T.; Brattain, M.G. DNA breakage in human lung carcinoma cells and nuclei that are naturally sensitive or resistant to etoposide and teniposide. Cancer Res. 1986, 46, 3809–3816. [Google Scholar] [PubMed]
- McMillan, E.A.; Ryu, M.-J.; Diep, C.H.; Mendiratta, S.; Clemenceau, J.; Vaden, R.M.; Kim, J.-H.; Motoyaji, T.; Covington, K.R.; Peyton, M.; et al. Chemistry-First Approach for Nomination of Personalized Treatment in Lung Cancer. Cell 2018, 173, 864–878.e29. [Google Scholar] [CrossRef] [Green Version]
- Suter, B.; Pogoutse, O.; Guo, X.; Krogan, N.; Lewis, P.; Greenblatt, J.F.; Rine, J.; Emili, A. Association with the origin recognition complex suggests a novel role for histone acetyltransferase Hat1p/Hat2p. BMC Biol. 2007, 5, 38. [Google Scholar] [CrossRef] [Green Version]
- Bulusu, V.; Tumanov, S.; Michalopoulou, E.; Van Den Broek, N.J.; MacKay, G.; Nixon, C.; Kamphorst, J.J. Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep. 2017, 18, 647–658. [Google Scholar] [CrossRef] [Green Version]
- Comerford, S.A.; Huang, Z.; Du, X.; Wang, Y.; Cai, L.; Witkiewicz, A.K.; Walters, H.; Tantawy, M.N.; Fu, A.; Manning, H.C.; et al. Acetate dependence of tumors. Cell 2014, 159, 1591–1602. [Google Scholar] [CrossRef] [Green Version]
- Kamphorst, J.J.; Chung, M.K.; Fan, J.; Rabinowitz, J.D. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab. 2014, 2, 23. [Google Scholar] [CrossRef]
- Schug, Z.T.; Peck, B.; Jones, D.T.; Zhang, Q.; Grosskurth, S.; Alam, I.S.; Goodwin, L.M.; Smethurst, E.; Mason, S.; Blyth, K.; et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 2015, 27, 57–71. [Google Scholar] [CrossRef] [Green Version]
- Garcia, P.A.A.; Hoover, M.E.; Zhang, P.; Nagarajan, P.; Freitas, M.A.; Parthun, M.R. Identification of multiple roles for histone acetyltransferase 1 in replication-coupled chromatin assembly. Nucleic Acids Res. 2017, 45, 9319–9335. [Google Scholar]
- Ge, Z.; Wang, H.; Parthun, M.R. Nuclear Hat1p complex (NuB4) components participate in DNA repair-linked chromatin reassembly. J. Biol. Chem. 2011, 286, 16790–16799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagarajan, P.; Ge, Z.; Sirbu, B.; Doughty, C.; Garcia, P.A.A.; Schlederer, M.; Annunziato, A.T.; Cortez, D.; Kenner, L.; Parthun, M.R. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet. 2013, 9, e1003518. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Parthun, M.R. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol. Cell. Biol. 2002, 22, 8353–8365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tscherner, M.; Stappler, E.; Hnisz, D.; Kuchler, K. The histone acetyltransferase Hat1 facilitates DNA damage repair and morphogenesis in Candida albicans. Mol. Microbiol. 2012, 86, 1197–1214. [Google Scholar] [CrossRef]
- Yang, X.; Li, L.; Liang, J.; Shi, L.; Yang, J.; Yi, X.; Zhang, D.; Han, X.; Yu, N.; Shang, Y. Histone acetyltransferase 1 promotes homologous recombination in DNA repair by facilitating histone turnover. J. Biol. Chem. 2013, 288, 18271–18282. [Google Scholar] [CrossRef] [Green Version]
- Garcia, P.A.A.; Lovejoy, C.M.; Nagarajan, P.; Park, D.; Popova, L.V.; Freitas, M.A.; Parthun, M.R. Histone acetyltransferase 1 is required for DNA replication fork function and stability. J. Biol. Chem. 2020, 295, 8363–8373. [Google Scholar] [CrossRef]
- Nagarajan, P.; Garcia, P.A.A.; Iyer, C.C.; Popova, L.V.; Arnold, W.D.; Parthun, M.R. Early-onset aging and mitochondrial defects associated with loss of histone acetyltransferase 1 (Hat1). Aging Cell 2019, 18, e12992. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Blank, M. Next-Generation Analysis of Deep Sequencing Data: Bringing Light into the Black Box of SELEX Experiments. Methods Mol. Biol. 2016, 1380, 85–95. [Google Scholar]
- Kwok, C.K.; Merrick, C.J. G-Quadruplexes: Prediction, Characterization, and Biological Application. Trends Biotechnol. 2017, 35, 997–1013. [Google Scholar] [CrossRef]
- Li, J.; Deng, H.; Hu, M.; Fang, Y.; Vaughn, A.; Cai, X.; Xu, L.; Wan, W.; Li, Z.; Chen, S.; et al. Inhibition of non-small cell lung cancer (NSCLC) growth by a novel small molecular inhibitor of EGFR. Oncotarget 2015, 6, 6749–6761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steen, N.V.; Potze, L.; Giovannetti, E.; Cavazzoni, A.; Ruijtenbeek, R.; Rolfo, C.; Pauwels, P.; Peters, G.J. Molecular mechanism underlying the pharmacological interactions of the protein kinase C-beta inhibitor enzastaurin and erlotinib in non-small cell lung cancer cells. Am. J. Cancer Res. 2017, 7, 816–830. [Google Scholar] [PubMed]
- Rusin, S.F.; Adamo, M.E.; Kettenbach, A.N. Identification of Candidate Casein Kinase 2 Substrates in Mitosis by Quantitative Phosphoproteomics. Front. Cell Dev. Biol. 2017, 5, 97. [Google Scholar] [CrossRef] [PubMed]
- Roffey, S.E.; Litchfield, D.W. CK2 Regulation: Perspectives in 2021. Biomedicines 2021, 9, 1361. [Google Scholar] [CrossRef]
- Yde, C.W.; Olsen, B.B.; Meek, D.; Watanabe, N.; Guerra, B. The regulatory beta-subunit of protein kinase CK2 regulates cell-cycle progression at the onset of mitosis. Oncogene 2008, 27, 4986–4997. [Google Scholar] [CrossRef] [PubMed]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klett-Mingo, J.I.; Pinto-Díez, C.; Cambronero-Plaza, J.; Carrión-Marchante, R.; Barragán-Usero, M.; Pérez-Morgado, M.I.; Rodríguez-Martín, E.; Toledo-Lobo, M.V.; González, V.M.; Martín, M.E. Potential Therapeutic Use of Aptamers against HAT1 in Lung Cancer. Cancers 2023, 15, 227. https://doi.org/10.3390/cancers15010227
Klett-Mingo JI, Pinto-Díez C, Cambronero-Plaza J, Carrión-Marchante R, Barragán-Usero M, Pérez-Morgado MI, Rodríguez-Martín E, Toledo-Lobo MV, González VM, Martín ME. Potential Therapeutic Use of Aptamers against HAT1 in Lung Cancer. Cancers. 2023; 15(1):227. https://doi.org/10.3390/cancers15010227
Chicago/Turabian StyleKlett-Mingo, José Ignacio, Celia Pinto-Díez, Julio Cambronero-Plaza, Rebeca Carrión-Marchante, Miriam Barragán-Usero, María Isabel Pérez-Morgado, Eulalia Rodríguez-Martín, Mª Val Toledo-Lobo, Víctor M González, and M. Elena Martín. 2023. "Potential Therapeutic Use of Aptamers against HAT1 in Lung Cancer" Cancers 15, no. 1: 227. https://doi.org/10.3390/cancers15010227
APA StyleKlett-Mingo, J. I., Pinto-Díez, C., Cambronero-Plaza, J., Carrión-Marchante, R., Barragán-Usero, M., Pérez-Morgado, M. I., Rodríguez-Martín, E., Toledo-Lobo, M. V., González, V. M., & Martín, M. E. (2023). Potential Therapeutic Use of Aptamers against HAT1 in Lung Cancer. Cancers, 15(1), 227. https://doi.org/10.3390/cancers15010227