Development and Validation of the Oxidative Stress Related lncRNAs for Prognosis in Esophageal Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
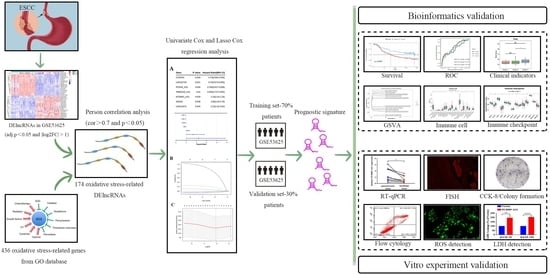
2. Materials and Methods
2.1. Data Source
2.2. Identification of DElncRNAs
2.3. Establishment and Validation of the Prognostic Model Using Oxidative Stress-Related DElncRNAs
2.4. Establishment and Validation of the Nomogram for the Prognostic Model
2.5. Gene Set Variation Analysis (GSVA) Analysis
2.6. Evaluation of Immune Infiltration and Immune Checkpoint Gene Expression between the High-Risk and Low-Risk Groups
2.7. Tissue Samples and ESCC Cell Lines
2.8. RNA Extraction and Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
2.9. Fluorescence In Situ Hybridization (FISH) Assay
2.10. Cell Transfection
2.11. Cell Viability Assay
2.12. Colony Formation Assay
2.13. Flow Cytometry Analysis
2.14. Reactive Oxygen Species (ROS) Detection
2.15. Determination of Lactate Dehydrogenase (LDH) Release
2.16. Statistical Analysis
3. Results
3.1. Screening Results of DElncRNAs in GSE53625 Dataset
3.2. Establishment and Validation of a Risk Model
3.3. Construction of the Nomogram
3.4. Association of the Risk Score and Clinicopathological Traits
3.5. GSVA between High-Risk and Low-Risk Groups
3.6. Correlations with Immune Microenvironment
3.7. The Oxidative Stress-Related DElncRNA Validation in ESCC
3.8. Loss of PCDH9-AS1 Predicted Unfavorable Prognosis of ESCC Patients
3.9. Overexpression of PCDH9-AS1 Attenuates ESCC Cell Proliferation and Promotes Apoptosis and Oxidative Stress Level
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, M.; Roshandel, G.; McCormack, V.; Malekzadeh, R. Current Status and Future Prospects for Esophageal Cancer. Cancers 2023, 15, 765. [Google Scholar] [CrossRef]
- Waters, J.K.; Reznik, S.I. Update on Management of Squamous Cell Esophageal Cancer. Curr. Oncol. Rep. 2022, 24, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.E.; Sewastjanow-Silva, M.; Waters, R.E.; Ajani, J.A. Esophageal cancer: Emerging therapeutics. Expert Opin. Ther. Targets 2022, 26, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Dimitrova, N.; Zamudio, J.R.; Jong, R.M.; Soukup, D.; Resnick, R.; Sarma, K.; Ward, A.J.; Raj, A.; Lee, J.T.; Sharp, P.A.; et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell 2014, 54, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Teng, L.; Feng, Y.C.; Guo, S.T.; Wang, P.L.; Qi, T.F.; Yue, Y.M.; Wang, S.X.; Zhang, S.N.; Tang, C.X.; La, T.; et al. The pan-cancer lncRNA PLANE regulates an alternative splicing program to promote cancer pathogenesis. Nat. Commun. 2021, 12, 3734–3750. [Google Scholar] [CrossRef]
- Lin, Q.Y.; Liu, T.; Wang, X.W.; Hou, G.X.; Xiang, Z.Y.; Zhang, W.X.; Zheng, S.L.; Zhao, D.; Leng, Q.B.; Zhang, X.S.; et al. Long noncoding RNA HITT coordinates with RGS2 to inhibit PD-L1 translation in T cell immunity. J. Clin. Investig. 2023, 133, e162951–e162971. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal. Transduct. Target. Ther. 2021, 6, 425–441. [Google Scholar] [CrossRef]
- Xue, S.T.; Zhang, B.; Cao, S.Q.; Ding, J.C.; Hu, G.S.; Liu, W.; Chen, C. Long non-coding RNA LINC00680 functions as a ceRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis. Mol. Cancer 2022, 21, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Wang, Z.; Shi, H.; Li, H.; Li, L.L.; Fang, R.P.; Cai, X.L.; Liu, B.W.; Zhang, X.D.; Ye, L.H. HBXIP and LSD1 Scaffolded by lncRNA Hotair Mediate Tranional Activation by c-Myc. Cancer Res. 2016, 76, 293–304. [Google Scholar] [CrossRef]
- Yang, X.; Zeng, T.Y.; Liu, Z.Y.; He, W.L.; Hu, M.T.; Tang, T.; Chen, L.; Xing, L. Long noncoding RNA GK-IT1 promotes esophageal squamous cell carcinoma by regulating MAPK1 phosphorylation. Cancer Med. 2022, 11, 4555–4574. [Google Scholar] [CrossRef]
- Luo, J.; Xie, K.; Gao, X.; Yao, Y.; Wang, G.M.; Shao, C.Y.; Li, X.K.; Xu, Y.; Ren, B.H.; Hu, L.W.; et al. Long Noncoding RNA Nuclear Paraspeckle Assembly Transcript 1 Promotes Progression and Angiogenesis of Esophageal Squamous Cell Carcinoma Through miR-590-3p/MDM2 Axis. Front. Oncol. 2021, 10, 618930–618941. [Google Scholar] [CrossRef]
- Chou, J.J.; Kaller, M.; Jaeckel, S.; Rokavec, M.; Hermeking, H. AP4 suppresses DNA damage, chromosomal instability and senescence via inducing MDC1/Mediator of DNA damage Checkpoint 1 and repressing MIR22HG/miR-22-3p. Mol. Cancer 2022, 21, 120–143. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.J.; Yang, Y.Q.; Li, M.Y.; Li, C.X.; Zhou, Z.T.; Tang, G.Y.; Wu, L.; Yao, Y.L.; Shen, X.M.; Hou, Z.Y.; et al. LncRNA IFITM4P promotes immune escape by up-regulating PD-L1 via dual mechanism in oral carcinogenesis. Mol. Ther. 2022, 30, 1564–1577. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Barwal, T.S.; Khandelwal, A.; Rana, M.K.; Rana, A.P.S.; Singh, K.; Jain, A. Circulating Long Non-Coding RNAs LINC00324 and LOC100507053 as Potential Liquid Biopsy Markers for Esophageal Squamous Cell Carcinoma: A Pilot Study. Front. Oncol. 2022, 12, 823953–823967. [Google Scholar] [CrossRef]
- Tang, J.Y.; Yang, O.; Hou, M.F.; Huang, H.W.; Wang, H.R.; Li, K.T.; Fayyaz, S.; Shu, C.W.; Chang, H.W. Oxidative stress-modulating drugs have preferential anticancer effects involving the regulation of apoptosis, DNA damage, endoplasmic reticulum stress, autophagy, metabolism, and migration. Semin. Cancer Biol. 2019, 58, 109–117. [Google Scholar] [CrossRef]
- Bai, R.; Guo, J.N.; Ye, X.Y.; Xie, Y.Y.; Xie, T. Oxidative stress: The core pathogenesis and mechanism of Alzheimer’s disease. Ageing Res. Rev. 2022, 77, 101619–101626. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Zhang, Q.; Zheng, R. The interplay between oxidative stress and autophagy in chronic obstructive pulmonary disease. Front. Physiol. 2022, 13, 1004275–1004289. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Zhang, G.Y.; Dasgupta, S.; Niewold, E.L.; Li, C.; Li, Q.F.; Luo, X.; Tan, L.; Ferdous, A.; Lorenzi, P.L.; et al. ATF4 Protects the Heart from Failure by Antagonizing Oxidative Stress. Circ. Res. 2022, 131, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yu, S.; Li, X.; Wen, X.; Liu, S.; Zu, R.; Ren, H.; Li, T.; Yang, C.; Luo, H. Research progress on the interaction between oxidative stress and platelets: Another avenue for cancer? Pharmacol. Res. 2023, 191, 106777–106788. [Google Scholar] [CrossRef]
- Liu, P.F.; Farooqi, A.A.; Peng, S.Y.; Yu, T.J.; Dahms, H.U.; Lee, C.H.; Tang, J.Y.; Wang, S.C.; Shu, C.W.; Chang, H.W. Regulatory effects of noncoding RNAs on the interplay of oxidative stress and autophagy in cancer malignancy and therapy. Semin. Cancer Biol. 2022, 83, 269–282. [Google Scholar] [CrossRef]
- Shi, X.B.; Li, Y.; Pan, S.P.; Liu, X.X.; Ke, Y.; Guo, W.; Wang, Y.C.; Ruan, Q.L.; Zhang, X.Z.; Ma, H.B. Identification and validation of an autophagy-related gene signature for predicting prognosis in patients with esophageal squamous cell carcinoma. Sci. Rep. 2022, 12, 1960–1973. [Google Scholar] [CrossRef]
- Zhao, M.N.; Jin, X.; Chen, Z.C.; Zhang, H.; Zhan, C.; Wang, H.; Wang, Q. Weighted Correlation Network Analysis of Cancer Stem Cell-Related Prognostic Biomarkers in Esophageal Squamous Cell Carcinoma. Technol. Cancer Res. Treat. 2022, 21, 15330338221117003–15330338221117011. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.F.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47–e59. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Morgan, E.; Soerjomataram, I.; Rumgay, H.; Coleman, H.G.; Thrift, A.P.; Vignat, J.; Laversanne, M.; Ferlay, J.; Arnold, M. The Global Landscape of Esophageal Squamous Cell Carcinoma and Esophageal Adenocarcinoma Incidence and Mortality in 2020 and Projections to 2040: New Estimates from GLOBOCAN 2020. Gastroenterology 2022, 163, 649–658.e2. [Google Scholar] [CrossRef]
- Chu, L.Y.; Peng, Y.H.; Weng, X.F.; Xie, J.J.; Xu, Y.W. Blood-based biomarkers for early detection of esophageal squamous cell carcinoma. World J. Gastroenterol. 2020, 26, 1708–1725. [Google Scholar] [CrossRef]
- Zhu, J.; Zhao, Y.; Wu, G.; Zhang, X.J.; Chen, Q.Q.; Yang, B.; Guo, X.W.; Ji, S.J.; Gu, K. Ferroptosis-Related lncRNA Signature Correlates with the Prognosis, Tumor Microenvironment, and Therapeutic Sensitivity of Esophageal Squamous Cell Carcinoma. Oxid. Med. Cell Longev. 2022, 2022, 7465880–7465907. [Google Scholar] [CrossRef]
- Sasagawa, S.; Kato, H.; Nagaoka, K.; Sun, C.B.; Imano, M.; Sato, T.; Johnson, T.A.; Fujita, M.; Maejima, K.; Okaw, Y.; et al. Mmuno-genomic profiling of biopsy specimens predicts neoadjuvant chemotherapy response in esophageal squamous cell carcinoma. Cell Rep. Med. 2022, 3, 100705–100721. [Google Scholar] [CrossRef]
- Ren, Q.H.; Zhang, P.P.; Zhang, X.; Feng, Y.L.; Li, L.; Lin, H.R.; Yu, Y. A fibroblast-associated signature predicts prognosis and immunotherapy in esophageal squamous cell cancer. Front. Immunol. 2023, 14, 1199040–1199055. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Zhang, J.H.; Xiu, Y.J.; Jin, Y.; Xiang, J.Q.; Nie, Y.Z.; Fu, S.; Zhao, K.L. Prognostic value of an immunohistochemical signature in patients with esophageal squamous cell carcinoma undergoing radical esophagectomy. Mol. Oncol. 2018, 12, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.P.; Song, X.; Zhao, X.K.; Wei, M.X.; Gao, S.G.; Zhou, F.Y.; Han, X.N.; Xu, R.H.; Wang, R.; Fan, Z.M.; et al. Serum Metabolomic Profiling Reveals Biomarkers for Early Detection and Prognosis of Esophageal Squamous Cell Carcinoma. Front. Oncol. 2022, 12, 790933–790946. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Li, L.; Feng, R.Y.; Li, Y.; Yu, X.; Liu, Y.H.; Zhao, Y.H.; Liu, Z.H. Caspase-8 mutants activate Nrf2 via phosphorylating SQSTM1 to protect against oxidative stress in esophageal squamous cell carcinoma. Free Radic. Biol. Med. 2022, 192, 51–62. [Google Scholar] [CrossRef]
- Liu, Z.C.; Gu, S.R.; Lu, T.C.; Wu, K.Q.; Li, L.; Dong, C.L.; Zhou, Y.X. IFI6 depletion inhibits esophageal squamous cell carcinoma progression through reactive oxygen species accumulation via mitochondrial dysfunction and endoplasmic reticulum stress. J. Exp. Clin. Cancer Res. 2020, 39, 144–171. [Google Scholar] [CrossRef]
- Liu, W.; Yang, H.S.; Zheng, S.Y.; Luo, H.H.; Feng, Y.F.; Lei, Y.Y. Oxidative stress genes in patients with esophageal squamous cell carcinoma: Construction of a novel prognostic signature and characterization of tumor microenvironment infiltration. BMC Bioinform. 2022, 23, 406–424. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Wang, J.; Sun, Y.; Zhang, X.; Cai, H.; Zhang, C.; Qu, H.; Liu, L.; Zhang, M.; Fu, J.; Zhang, J.; et al. Oxidative stress activates NORAD expression by H3K27ac and promotes oxaliplatin resistance in gastric cancer by enhancing autophagy flux via targeting the miR-433-3p. Cell Death Dis. 2021, 12, 90–104. [Google Scholar] [CrossRef]
- Wang, W.T.; Ye, H.; Wei, P.P.; Han, B.W.; He, B.; Chen, Z.H.; Chen, Y.Q. LncRNAs H19 and HULC, activated by oxidative stress, promote cell migration and invasion in cholangiocarcinoma through a ceRNA manner. J. Hematol. Oncol. 2016, 9, 117–129. [Google Scholar] [CrossRef]
- Ma, L.Y.; Xu, A.; Kang, L.; Cong, R.; Fan, Z.Y.; Zhu, X.; Huo, N.; Liu, W.P.; Xue, C.Y.; Ji, Q.B.; et al. LSD1-Demethylated LINC01134 Confers Oxaliplatin Resistance Through SP1-Induced p62 Transcription in HCC. Hepatology 2021, 74, 3213–3234. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Li, Z.; Xia, L.; Dai, J.; Zhang, Q.; Wu, C.; Xu, S. LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress during erastin-induced ferroptosis in HepG2 hepatocellular carcinoma cells. Sci. Rep. 2019, 9, 16185–16196. [Google Scholar] [CrossRef] [PubMed]
- D′Souza, L.C.; Mishra, S.; Chakraborty, A.; Shekher, A.; Sharma, A.; Gupta, S.C. Oxidative Stress and Cancer Development: Are Noncoding RNAs the Missing Links? Antioxid. Redox Signal. 2020, 33, 1209–1229. [Google Scholar] [CrossRef]
- Wu, Y.Z.; Su, Y.H.; Kuo, C.Y. Stressing the Regulatory Role of Long Non-Coding RNA in the Cellular Stress Response during Cancer Progression and Therapy. Biomedicines 2022, 10, 1212. [Google Scholar] [CrossRef] [PubMed]
- Li, F.W.; Luo, S.K. Identification and Construction of a Predictive Immune-Related lncRNA Signature Model for Melanoma. Int. J. Gen. Med. 2021, 14, 9227–9235. [Google Scholar] [CrossRef]
- Zhang, J.F.; Ling, X.D.; Fang, C.Y.; Ma, J.Q. Identification and validation of an eight-lncRNA signature that predicts prognosis in patients with esophageal squamous cell carcinoma. Cell Mol. Biol. Lett. 2022, 27, 39–56. [Google Scholar] [CrossRef]
- Cao, C.; Li, J.; Li, G.; Hu, G.; Deng, Z.; Huang, B.; Yang, J.; Li, J.; Cao, S. Long Non-coding RNA TMEM220-AS1 Suppressed Hepatocellular Carcinoma by Regulating the miR-484/MAGI1 Axis as a Competing Endogenous RNA. Front. Cell Dev. Biol. 2021, 9, 681529–681541. [Google Scholar] [CrossRef]
- Ma, S.Y.; Wei, P.; Qu, F. KCNMA1-AS1 attenuates apoptosis of epithelial ovarian cancer cells and serves as a risk factor for poor prognosis of epithelial ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4629–4641. [Google Scholar] [CrossRef]
- Zhang, Y.; Jin, X.; Wang, Z.; Zhang, X.; Liu, S.; Liu, G. Downregulation of SNHG1 suppresses cell proliferation and invasion by regulating Notch signaling pathway in esophageal squamous cell cancer. Cancer Biomark. 2017, 21, 89–96. [Google Scholar] [CrossRef]
- Cai, H.; Xu, H.; Lu, H.; Xu, W.; Liu, H.; Wang, X.; Zhou, G.; Yang, X. LncRNA SNHG1 Facilitates Tumor Proliferation and Represses Apoptosis by Regulating PPARγ Ubiquitination in Bladder Cancer. Cancers 2022, 14, 4740. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.G.; Luo, M.; Zhou, C.C.; Shi, X.J.; Yang, W.H.; Lu, Z.L.; Chen, Z.L.; Sun, N.; He, J. Novel long noncoding RNA NMR promotes tumor progression via NSUN2 and BPTF in esophageal squamous cell carcinoma. Cancer Lett. 2018, 430, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Jia, Y.; Wang, J.; Liu, T.; Cheng, Z.; Sang, M.; Lv, W.; Qin, J.; Liu, L. Long noncoding RNA DGCR5 involves in tumorigenesis of esophageal squamous cell carcinoma via SRSF1-mediated alternative splicing of Mcl-1. Cell Death Dis. 2021, 12, 587–600. [Google Scholar] [CrossRef]
- Li, H.; An, J.; Wu, M.; Zheng, Q.; Gui, X.; Li, T.; Pu, H.; Lu, D. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget 2015, 6, 27847–27864. [Google Scholar] [CrossRef] [PubMed]
- Rebernick, R.J.; Bell, H.N.; Bauer, T.M.; McEwen, D.; Werkman, D.F.; Chang, A.C.; Lin, J.; Reddy, R.M.; Kresty, L.; Lagisetty, K. Role of IL4 and GMCSF in Predicting Survival in Esophageal Cancer. J. Am. Coll. Surg. 2023, 236, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Cai, L.; Xiong, P.; Liu, Z.; Chen, S.; Liu, X.; Lin, R.; Lei, Z.; Tian, D.; Su, M. TLR3 Expression is a Potential Prognosis Biomarker and Shapes the Immune-Active Tumor Microenvironment in Esophageal Squamous Cell Carcinoma. J. Inflamm. Res. 2022, 15, 1437–1456. [Google Scholar] [CrossRef]
- Zhao, Z.H.; Wang, Y.; Gao, Y.H.; Ju, Y.R.; Zhao, Y.; Wu, Z.F.; Gao, S.X.; Zhang, B.Y.; Pang, X.W.; Zhang, Y.; et al. The PRAK-NRF2 axis promotes the differentiation of Th17 cells by mediating the redox homeostasis and glycolysis. Proc. Natl. Acad. Sci. USA 2023, 120, e2212613120–e2212613129. [Google Scholar] [CrossRef]
- Habtetsion, T.; Ding, Z.C.; Pi, W.H.; Li, T.; Lu, C.W.; Chen, T.T.; Xi, C.X.; Spartz, H.; Liu, K.B.; Hao, Z.L.; et al. Alteration of Tumor Metabolism by CD4+ T Cells Leads to TNF-α-Dependent Intensification of Oxidative Stress and Tumor Cell Death. Cell Metab. 2018, 28, 228–242.e6. [Google Scholar] [CrossRef]
- Zhuge, L.; Huang, B.; Xie, J.; Gao, Z.; Zheng, D.; Zheng, S.; Xiang, J.; Zhang, J. Immunoscore Signature Predicts Postoperative Survival and Adjuvant Chemotherapeutic Benefits in Esophageal Squamous Cell Carcinoma. Cancer Manag. Res. 2020, 12, 12885–12894. [Google Scholar] [CrossRef]
- Wang, J.B.; Jia, Y.B.; Wang, N.N.; Zhang, X.M.; Tan, B.X.; Zhang, G.Y.; Cheng, Y.F. The clinical significance of tumor-infiltrating neutrophils and neutrophil-to-CD8+ lymphocyte ratio in patients with resectable esophageal squamous cell carcinoma. J. Transl. Med. 2014, 12, 7–16. [Google Scholar] [CrossRef]
- Mao, J.T.; Lu, Q.; Jing, P.Y.; Li, Z.L.; Yang, X.Q.; Zhang, J.P.; Li, Z. Comprehensive Analysis of Prognostic Value and Immune Infiltration of MMP12 in Esophageal Squamous Cell Carcinoma. J. Oncol. 2022, 2022, 4097428–4097438. [Google Scholar] [CrossRef]
- Que, H.; Fu, Q.; Lan, T.; Tian, X.; Wei, X. Tumor-associated neutrophils and neutrophil-targeted cancer therapies. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188762–188879. [Google Scholar] [CrossRef]
- Komi, D.E.A.; Redegeld, F.A. Role of Mast Cells in Shaping the Tumor Microenvironment. Clin. Rev. Allergy Immunol. 2020, 58, 313–325. [Google Scholar] [CrossRef]










| Targets | Forward Sequence (5′→3′) | Reverse Sequence (5′→3′) |
|---|---|---|
| CCR5AS | AACATTTGGTGCCGAAGACC | CATGGAGTGAGGGTGAGGAG |
| LINC01749 | GGCCTCTCTTGAAGGGACTT | GGCCTGACACACGAATGTTT |
| PCDH9-AS1 | TTTAGGAAAGGAACTATTATCAC | GCTTATTATTGCCTATAAACGAC |
| TMEM220-AS1 | AGGGAGCCACTCTGCCCTTGTTT | ATGAGGACTGTGAAGCCGAGAAA |
| KCNMA1-AS1 | F: GGGACATTGGGAGGAACAGA | ACCAGCAGGGCTAATAGCAG |
| SNHG1 | F: CCTGCAAGCCTCTTGCTTAG | TGGGCTGAACATTGCAACAA |
| LINC01672 | F: GGCAAAAACCAGGAGATCCCA | GCCATGTCATTAGCCACCAG |
| GAPDH | F: ACCCACTCCTCCACCTTTGA | CCACCCTGTTGCTGTAGCCA |
| Characteristic | n | PCDH9-AS1 Level | p-Value | |
|---|---|---|---|---|
| Low (n = 40) | High (n = 40) | |||
| Gender | ||||
| Male | 56 | 30 | 26 | 0.3291 |
| Female | 24 | 10 | 14 | |
| Age (years) # | ||||
| ≤60 | 33 | 19 | 14 | 0.2958 |
| >60 | 46 | 21 | 25 | |
| Histological grade | ||||
| I + II | 65 | 34 | 31 | 0.3902 |
| III + IV | 15 | 6 | 9 | |
| Lymph node metastasis | ||||
| Negative | 41 | 18 | 23 | 0.2634 |
| Positive | 39 | 22 | 17 | |
| Tumor Stage # | ||||
| T1 + T2 | 9 | 1 | 8 | 0.0714 |
| T3 + T4 | 27 | 12 | 15 | |
| Clinical stage # | ||||
| I + II | 19 | 4 | 15 | 0.0258 |
| III + IV | 28 | 15 | 13 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Liu, W.; Zhu, Y.; Kong, W.; Su, X.; Huang, L.; Cui, Y.; Sun, G. Development and Validation of the Oxidative Stress Related lncRNAs for Prognosis in Esophageal Squamous Cell Carcinoma. Cancers 2023, 15, 4399. https://doi.org/10.3390/cancers15174399
Zheng X, Liu W, Zhu Y, Kong W, Su X, Huang L, Cui Y, Sun G. Development and Validation of the Oxidative Stress Related lncRNAs for Prognosis in Esophageal Squamous Cell Carcinoma. Cancers. 2023; 15(17):4399. https://doi.org/10.3390/cancers15174399
Chicago/Turabian StyleZheng, Xuan, Wei Liu, Yingze Zhu, Wenyue Kong, Xin Su, Lanxiang Huang, Yishuang Cui, and Guogui Sun. 2023. "Development and Validation of the Oxidative Stress Related lncRNAs for Prognosis in Esophageal Squamous Cell Carcinoma" Cancers 15, no. 17: 4399. https://doi.org/10.3390/cancers15174399
APA StyleZheng, X., Liu, W., Zhu, Y., Kong, W., Su, X., Huang, L., Cui, Y., & Sun, G. (2023). Development and Validation of the Oxidative Stress Related lncRNAs for Prognosis in Esophageal Squamous Cell Carcinoma. Cancers, 15(17), 4399. https://doi.org/10.3390/cancers15174399