A Transfer-Learning-Based Novel Convolution Neural Network for Melanoma Classification
Abstract
:1. Introduction
2. Material and Methods
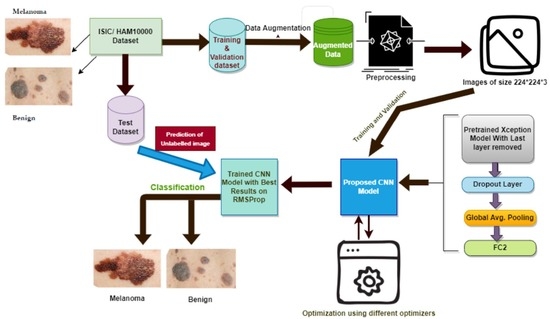
- Dataset selection and augmentation: ISIC archive dataset is selected, and data augmentation is applied to overcome the class imbalance problem, leading to overfitting;
- Preprocessing: Input image preprocessing is kept minimum to increase the generalization ability and adaptability of the proposed model for other classification tasks;
- Selection of base model: Xception model [23], a well-known CNN model pre-trained on the ImageNet dataset, is selected as the base model;
- The last layer of the Xception model is removed, and three more layers are added: drop out layer, global average pooling, and dense or FC layer with two nodes, which is the last layer for binary classification, i.e., melanoma or benign;
- The pre-trained Xception network is fine-tuned multiple times on the ISIC archive dataset with different settings to achieve more accuracy and good performance for skin lesion classification;
- The model is optimized using six different optimizers, and the model performance is compared on each of those optimizers;
- A novel variable learning rate algorithm and early stopping criterion are used to avoid unnecessary model training for fix number of epochs;
- Results: The proposed method’s performance in every different setting is shown and compared to other techniques developed so far.
- Figure 1 below shows the steps that are followed in the proposed methodology.
2.1. Dataset and Its Augmentation
2.2. Preprocessing
2.3. Pre-Trained Convolution Neural Network Model and Its Fine-Tuning
3. Experimental Setup and Results
3.1. Training and Validation Process
3.2. Testing Process
4. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNN | Convolution Neural Network |
| ISIC | International Skin Image Collaboration |
| BRAF | Human Gene That Encodes a Protein Called B-Raf |
| VGG | Visual Geometry Group |
| ResNet | Residual Network |
| Adam | Adaptive Moment Estimation |
| Nadam | Nesterov-Accelerated Adaptive Moment Estimation |
| SGD | Stochastic Gradient Descent |
| RMSProp | Root Mean Square Propagation |
| GPU | Graphics Processing Unit |
| TPU | Tensor Processing Unit |
| ROC | Receiver Operating Characteristic Curve |
| AUC | Area Under ROC Curve |
References
- American Cancer Society: Melanoma Skin Cancer. Available online: https://www.cancer.org/cancer/melanoma-skin-cancer/ (accessed on 9 April 2022).
- Goncharova, Y.; Attia, E.A.; Souid, K.; Vasilenko, I.V. Dermoscopic features of facial pigmented skin lesions. ISRN Dermatol. 2013, 2013, 546813. [Google Scholar] [CrossRef]
- Zalaudek, I.; Lallas, A.; Moscarella, E.; Longo, C.; Soyer, H.P.; Argenziano, G. The dermatologist’s stethoscope-traditional and new applications of dermoscopy. Dermatol. Pract. Concept. 2013, 3, 67–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argenziano, G.; Soyer, H.P. Dermoscopy of pigmented skin lesions—A valuable tool for early diagnosis of melanoma. Lancet Oncol. 2001, 2, 443–449. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Gola Isasi, A.; García Zapirain, B.; Méndez Zorrilla, A. Melanomas non-invasive diagnosis application based on the ABCD rule and pattern recognition image processing algorithms. Comput. Biol. Med. 2011, 41, 742–755. [Google Scholar] [CrossRef] [PubMed]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the 25th International Conference on Neural Information Processing Systems—Volume 1 (NIPS’12), Red Hook, NY, USA, 3–6 December 2012; pp. 1097–1105. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations (ICLR2015), San Diego, CA, USA, 7–9 May 2015; Available online: https://arxiv.org/abs/1409.1556 (accessed on 10 November 2021).
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Miami, FL, USA, 20–25 June 2009. [Google Scholar]
- Vasconcelos, C.; Vasconcelos, B.N. Increasing Deep Learning Melanoma Classification by Classical and Expert Knowledge Based Image Transforms. arXiv 2017, arXiv:abs/1702.07025. [Google Scholar]
- Lopez, A.R.; Giro-i Nieto, X.; Burdick, J.; Marques, O. Skin lesion classification from dermoscopic images using deep learning techniques. In Proceedings of the 13th IASTED International Conference on Biomedical Engineering (BioMed), Innsbruck, Austria, 20–21 February 2017. [Google Scholar]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawahara, J.; BenTaieb, A.; Hamarneh, G. Deep features to classify skin lesions. In Proceedings of the 13th International Symposium on Biomedical Imaging, (IEEE), Prague, Czech Republic, 16 June 2016; pp. 1397–1400. [Google Scholar] [CrossRef]
- Codella, N.; Cai, J.; Abedini, M.; Garnavi, R.; Halpern, A.; Smith, J.R. Deep learning, sparse coding, and SVM for melanoma recognition in dermoscopy images. In Proceedings of the 6th International Workshop on Machine Learning in Medical Imaging, Held in Conjunction with MICCAI 2015, Munich, Germany, 5 October 2015; pp. 118–126. [Google Scholar]
- DeVries, T.; Ramachandram, D. Skin Lesion Classification Using Deep Multi-scale Convolutional Neural Networks. arXiv 2017, arXiv:1703.01402. [Google Scholar] [CrossRef]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. In Proceedings of the AAAI Conference on Artificial Intelligence, Stanford, CA, USA, 21–23 March 2016. [Google Scholar]
- Sabbaghi, S.; Aldeen, M.; Garnavi, R. A deep bag-of-features model for the classification of melanomas in dermoscopy images, 2016. In Proceedings of the 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 1369–1372. [Google Scholar] [CrossRef]
- Deivanayagampillai, N.; Suruliandi, A.; Kavitha, J. Melanoma Detection in Dermoscopic Images using Global and Local Feature Extraction. Int. J. Multimed. Ubiquitous Eng. 2017, 12, 19–27. [Google Scholar] [CrossRef]
- Nasiri, S.; Helsper, J.; Jung, M.; Fathi, M. DePicT Melanoma Deep-CLASS: A deep convolutional neural networks approach to classify skin lesion images. BMC Bioinform. 2020, 21 (Suppl. 2), 84. [Google Scholar] [CrossRef] [Green Version]
- Chollet, F. Xception: Deep Learning with Depth Wise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar]
- The International Skin Imaging Collaboration (ISIC). Available online: https://www.isic-archive.com/#!/topWithHeader/onlyHeaderTop/gallery (accessed on 20 November 2021).
- Mikołajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the 2018 International Interdisciplinary PhD Workshop (IIPhDW), Świnouście, Poland, 9–12 May 2018; pp. 117–122. [Google Scholar]
- Gu, S.; Pednekar, M.; Slater, R. Improve Image Classification Using Data Augmentation and Neural Networks. SMU Data Sci. Rev. 2019, 2, 1. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- LazyAdam a Variant of Adam Optimizer. Available online: https://tensorflow.org/addons/api_docs/python/tfa//optimizers//La-zyAdam (accessed on 18 December 2021).
- Dozat, T. Incorporating Nesterov Momentum into Adam. In Proceedings of the ICLR Workshop, San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Tieleman, T.; Hinton, G. Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude. Coursera Neural Netw. Mach. Learn. 2012, 4, 26–31. [Google Scholar]
- Robbins, H.; Monro, S. A stochastic approximation method. Ann. Math. Stat. 1951, 22, 400–407. [Google Scholar] [CrossRef]
- Prechelt, L. Early Stopping—But When? In Neural Networks: Tricks of the Trade; Montavon, G., Orr, G.B., Müller, K.R., Eds.; Lecture Notes in Computer Science; Springer: Berlin, Heidelberg, 2012; Volume 7700. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.E.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Nwankpa, C.; Ijomah, W.; Gachagan, A.; Marshall, S. Activation Functions: Comparison of trends in Practice and Research for Deep Learning. In Proceedings of the 2nd International Conference on Computational Sciences and Technology, Jamshoro, Pakistan, 17–19 December 2020. [Google Scholar]
- Harangi, B. Skin lesion classification with ensembles of deep convolutional neural networks. J. Biomed. Inform. 2018, 86, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Automated Melanoma Recognition in Dermoscopy Images via Very Deep Residual Networks. IEEE Trans. Med. Imaging 2017, 36, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Albahar, M.A. Skin Lesion Classification Using Convolutional Neural Network with Novel Regularizer. IEEE Access 2019, 7, 38306–38313. [Google Scholar] [CrossRef]
- Yoshida, T.; Celebi, M.E.; Schaefer, G.; Iyatomi, H. Simple and effective preprocessing for automated melanoma discrimination based on cytological findings. In Proceedings of the IEEE International Conference on Big Data, Washington, DC, USA, 5–8 December 2016. [Google Scholar]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-sources dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Sipon Miah, J.H.; Rahman, M.; Islam, K. An enhanced technique of skin cancer classification using deep convolutional neural network with transfer learning models. Mach. Learn. Appl. 2021, 5, 100036. [Google Scholar] [CrossRef]
- Sinha, D.; El-Sharkawy, M. Thin MobileNet: An Enhanced MobileNet Architecture. In Proceedings of the IEEE 10th Annual Ubiquitous Computing, Electronics & Mobile Communication Conference (UEMCON), New York, NY, USA, 10–12 October 2019; pp. 280–285. [Google Scholar] [CrossRef]
- Carcagnì, P.; Leo, M.; Cuna, A.; Mazzeo, P.L.; Spagnolo, P.; Celeste, G.; Distante, C. Classification of skin lesions by combining multilevel learnings in a densenet architecture. In Proceedings of the International Conference on Image Analysis and Processing, Trento, Italy, 9–13 September 2019; pp. 335–344. [Google Scholar] [CrossRef]
- Ameri, A. A Deep Learning Approach to Skin Cancer Detection in Dermoscopy Images. J. Biomed. Phys. Eng. 2020, 10, 801–806. [Google Scholar] [CrossRef]
- Pham, T.C.; Tran, G.S.; Nghiem, T.P.; Doucet, A.; Luong, C.M.; Hoang, V. A Comparative Study for Classification of Skin Cancer. In Proceedings of the International Conference on System Science and Engineering (ICSSE), Dong Hoi City, Vietnam, 19–21 July 2019. [Google Scholar] [CrossRef]
- Nugroho, A.; Slamet, I.; Sugiyanto. Skins cancer identification system of HAMl0000 skin cancer dataset using convolutional neural network. In Proceedings of the AIP Conference, Surakata, Indonesia, 19 July 2019; Volume 2202, p. 020039. [Google Scholar] [CrossRef]










| Optimizer | Training Loss | Training Accuracy | Validation Loss | Validation Accuracy | Total Epochs Reached | Training Time (in Seconds) |
|---|---|---|---|---|---|---|
| SGD | 0.2423 | 93.33% | 0.2059 | 95.38% | 66 | 9738.66 |
| Adam | 0.0060 | 99.78% | 0.0879 | 97.64% | 11 | 2170.34 |
| Adamax | 0.0478 | 98.25% | 0.0982 | 95.85% | 9 | 1345.32 |
| Nadam | 0.0364 | 98.59% | 0.1181 | 96.39% | 7 | 1092.50 |
| LazyAdam | 0.0073 | 99.83% | 0.0860 | 97.50% | 7 | 1128.70 |
| RMSProp | 0.0258 | 99.15% | 0.1431 | 97.94% | 10 | 1507.78 |
| Metrics | Optimizer | |||||
|---|---|---|---|---|---|---|
| SGD | Adam | Adamax | Nadam | LazyAdam | RMSProp | |
| Sparse_cc_loss | 0.229 | 0.142 | 0.106 | 0.228 | 0.132 | 0.225 |
| Benign_accuracy | 93.8% | 96.2% | 95.6% | 94.9% | 96.0% | 96.9% |
| Malignant_accuracy | 93.8% | 96.2% | 95.6% | 94.9% | 96.0% | 96.9% |
| Avg_accuracy | 93.8% | 96.2% | 95.6% | 94.9% | 96.0% | 96.8% |
| Avg_precision | 82.7% | 95.6% | 94.8% | 93.6% | 95.2% | 96.3% |
| Avg_specificity | 82.7% | 95.6% | 94.7% | 93.6% | 95.0% | 96.2% |
| Avg_sensitivity | 82.7% | 95.6% | 94.7% | 93.6% | 95.0% | 96.2% |
| Avg_AUC | 96.9% | 98.6% | 99.1% | 98.7% | 98.9% | 98.5% |
| Time taken (in secs) | 02 | 04 | 03 | 02 | 02 | 02 |
| Methods | Metrics | |||
|---|---|---|---|---|
| Accuracy | AUC | Sensitivity | Specificity | |
| An ensemble-based CNN framework [35] | 86.6% | 89% | 55.6% | 78.5% |
| FCRN melanoma recognition [36] | 85.5% | 78% | 54.7% | 93.1% |
| Novel regularizer-based CNN for skin lesion classification [37] | - | 98% | 94.3% | 93.6% |
| CNN model with cytological findings [38] | - | 84% | 80.9% | 88.1% |
| Proposed model | 96.8% | 99% | 96.2% | 96.2% |
| Model | Training Accuracy | Classification Accuracy | AUC |
|---|---|---|---|
| Proposed model | 98.81% | 91.54% | 86.41% |
| DCNN model [40] | 93.16% | 90.16% | NA |
| MobileNet [41] | 92.93% | 82.62% | NA |
| DenseNet [42] | 91.36% | 85.25% | NA |
| DLSC model [43] | NA | 84% | 91% |
| Comparative study [44] | NA | 74.75% | 81.46% |
| SCIS [45] | 80% | 78% | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qureshi, M.N.; Umar, M.S.; Shahab, S. A Transfer-Learning-Based Novel Convolution Neural Network for Melanoma Classification. Computers 2022, 11, 64. https://doi.org/10.3390/computers11050064
Qureshi MN, Umar MS, Shahab S. A Transfer-Learning-Based Novel Convolution Neural Network for Melanoma Classification. Computers. 2022; 11(5):64. https://doi.org/10.3390/computers11050064
Chicago/Turabian StyleQureshi, Mohammad Naved, Mohammad Sarosh Umar, and Sana Shahab. 2022. "A Transfer-Learning-Based Novel Convolution Neural Network for Melanoma Classification" Computers 11, no. 5: 64. https://doi.org/10.3390/computers11050064
APA StyleQureshi, M. N., Umar, M. S., & Shahab, S. (2022). A Transfer-Learning-Based Novel Convolution Neural Network for Melanoma Classification. Computers, 11(5), 64. https://doi.org/10.3390/computers11050064