Catalytic Biomaterials for Atrazine Degradation
Abstract
:1. Introduction
2. Results and Discussion
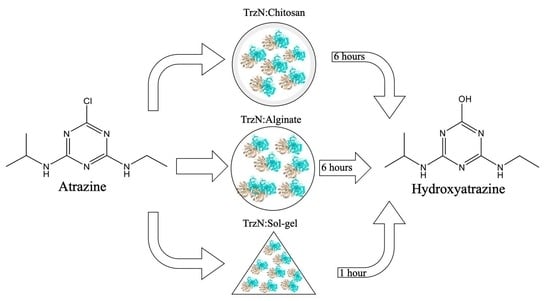
2.1. Encapsulation of TrzN
2.2. Proteolytic Digestion of WT TrzN and the TrzN:alginate, TrzN:chitosan, and TrzN:sol–gel Biomaterials
2.3. Thermostability of WT TrzN and the TrzN:alginate, TrzN:chitosan, and TrzN:sol–gel Biomaterials
2.4. Reusability and Long-Term Stability of the TrzN:biomaterials
2.5. Stability of the WT TrzN and TrzN:biomaterials in an Organic Co-Solvent
2.6. Stability of the WT TrzN and TrzN:biomaterials at Non-physiological pH Values
3. Materials and Methods
3.1. Materials
3.2. Expression and Purification of TrzN
3.3. Kinetic Activity Assay
3.4. Immobilization of TrzN in a Sol–Gel and Alginate Beads in the Absence and Presence of a Chitosan Coating
3.5. Kinetic characterization of the TrzN:Alginate, TrzN:Chitosan, and TrzN:sol–gel biomaterials
3.6. Proteolytic Digestion of Soluble TrzN, TrzN:alginate, TrzN:chitosan, and TrzN:sol–gel
3.7. Recycling Experiments for TrzN:alginate, TrzN:chitosan and TrzN:sol–gel
3.8. Activity of Soluble and Immobilized TrzN in Organic Co-Solvents
3.9. Activity of Soluble and Immobilized TrzN at Varying pH Values
3.10. Thermostability of Soluble and Immobilized TrzN
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pathak, R.K.; Dikshit, A.K. Atrazine and Human Health. Int. J. Ecosyst. 2012, 1, 14–23. [Google Scholar] [CrossRef] [Green Version]
- de Albuquerque, F.P.; de Oliveira, J.L.; Moschini-Carlos, V.; Fraceto, L.F. An Overview of the Potential Impacts of Atrazine in Aquatic Environments: Perspectives for Tailored Solutions Based on Nanotechnology. Sci. Total Environ. 2020, 700, 134868. [Google Scholar] [CrossRef] [PubMed]
- Solomon, K.R.; Baker, D.B.; Richards, R.P.; Dixon, K.R.; Klaine, S.J.; Point, T.W.L.; Kendall, R.J.; Weisskopf, C.P.; Giddings, J.M.; Giesy, J.P.; et al. Ecological Risk Assessment Of Atrazine In. Environ. Toxicol. 1996, 15, 31–76. [Google Scholar] [CrossRef]
- Beaulieu, M.; Cabana, H.; Taranu, Z.; Huot, Y. Predicting Atrazine Concentrations in Waterbodies across the Contiguous United States: The Importance of Land Use, Hydrology, and Water Physicochemistry. Limnol. Oceanogr. 2020, 65, 2966–2983. [Google Scholar] [CrossRef]
- Graymore, M.; Stagnitti, F.; Allinson, G. Impacts of Atrazine in Aquatic Ecosystems. Environ. Int. 2001, 26, 483–495. [Google Scholar] [CrossRef]
- Mudhoo, A.; Garg, V.K. Sorption, Transport and Transformation of Atrazine in Soils, Minerals and Composts: A Review. Pedosphere 2011, 21, 11–25. [Google Scholar] [CrossRef]
- Chernyak, S.M.; Rice, C.P.; McConnell, L.L. Evidence of Currently-Used Pesticides in Air, Ice, Fog, Seawater and Surface Microlayer in the Bering and Chukchi Seas. Mar. Pollut. Bull. 1996, 32, 410–419. [Google Scholar] [CrossRef]
- Ocking, D.A.J.H.; Abbitt, K.I.J.B. Amphibian Contributions To Ecosystem Services. Herpetol. Conserv. Biol. 2014, 9, 1–17. [Google Scholar]
- Hayes, T.B.; Khoury, V.; Narayan, A.; Nazir, M.; Parka, A.; Brown, T.; Adame, L.; Chan, E.; Buchholz, D.; Stueve, T.; et al. Atrazine Induces Complete Feminization and Chemical Castration in Male African Clawed Frogs (Xenopus Laevis). Proc. Natl. Acad. Sci. USA 2010, 107, 4612–4617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Wang, Y.; Zhu, Z.; Yang, E.; Feng, X.; Fu, Z.; Jin, Y. Atrazine and Its Main Metabolites Alter the Locomotor Activity of Larval Zebrafish (Danio Rerio). Chemosphere 2016, 148, 163–170. [Google Scholar] [CrossRef]
- ATSDR. Public Health Statement Atrazine; Agency for Toxic Substances and Disease Registry: Atlanta, GA, USA, 2015; pp. 5–8. [Google Scholar]
- Rostami, S.; Jafari, S.; Moeini, Z.; Jaskulak, M.; Keshtgar, L.; Badeenezhad, A.; Azhdarpoor, A.; Rostami, M.; Zorena, K.; Dehghani, M. Current Methods and Technologies for Degradation of Atrazine in Contaminated Soil and Water: A Review. Environ. Technol. Innov. 2021, 24, 102019. [Google Scholar] [CrossRef]
- Shapir, N.; Pedersen, C.; Gil, O.; Strong, L.; Seffernick, J.; Sadowsky, M.J.; Wackett, L.P. TrzN from Arthrobacter Aurescens TC1 Is a Zinc Amidohydrolase. J. Bacteriol. 2006, 188, 5859–5864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, Y.; Zhan, G.; Huang, C.; Wang, X.; Ai, Z.; Zou, J.; Luo, S.; Zhang, L. Dechlorination-Hydroxylation of Atrazine to Hydroxyatrazine with Thiosulfate: A Detoxification Strategy in Seconds. Environ. Sci. Technol. 2019, 53, 3208–3216. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. Enzyme Immobilization: The Quest for Optimum Performance. Adv. Synth. Catal. 2007, 349, 1289–1307. [Google Scholar] [CrossRef]
- Martinez, S.; Kuhn, M.L.; Russell, J.T.; Holz, R.C.; Elgren, T.E. Acrylamide Production Using Encapsulated Nitrile Hydratase from Pseudonocardia Thermophila in a Sol-Gel Matrix. J. Mol. Catal. B Enzym. 2014, 100, 19–24. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, X.; Jiang, Z.; Cao, B.; Ge, S.; Hu, M. Assessment of the Ecological Security of Immobilized Enzyme Remediation Process with Biological Indicators of Soil Health. Environ. Sci. Pollut. Res. 2013, 20, 5773–5780. [Google Scholar] [CrossRef]
- LI, Q.; LI, Y.; ZHU, X.; CAI, B. Isolation and Characterization of Atrazine-Degrading Arthrobacter Sp. AD26 and Use of This Strain in Bioremediation of Contaminated Soil. J. Environ. Sci. 2008, 20, 1226–1230. [Google Scholar] [CrossRef]
- Jackson, C.J.; Coppin, C.W.; Carr, P.D.; Aleksandrov, A.; Wilding, M.; Sugrue, E.; Ubels, J.; Paks, M.; Newman, J.; Peat, T.S.; et al. 300-Fold Increase in Production of the Zn2+-Dependent Dechlorinase TrzN in Soluble Form via Apoenzyme Stabilization. Appl. Environ. Microbiol. 2014, 80, 4003–4011. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Jeong, C.; Cho, S.; Kim, S.B. Effects of Thermal Treatment on the Physical Properties of Edible Calcium Alginate Gel Beads: Response Surface Methodological Approach. Foods 2019, 8, 578. [Google Scholar] [CrossRef] [Green Version]
- Fareez, I.M.; Lim, S.M.; Mishra, R.K.; Ramasamy, K. Chitosan Coated Alginate-Xanthan Gum Bead Enhanced PH and Thermotolerance of Lactobacillus Plantarum LAB12. Int. J. Biol. Macromol. 2015, 72, 1419–1428. [Google Scholar] [CrossRef]
- Won, K.; Kim, S.; Kim, K.J.; Park, H.W.; Moon, S.J. Optimization of Lipase Entrapment in Ca-Alginate Gel Beads. Process Biochem. 2005, 40, 2149–2154. [Google Scholar] [CrossRef]
- Bedade, D.K.; Sutar, Y.B.; Singhal, R.S. Chitosan Coated Calcium Alginate Beads for Covalent Immobilization of Acrylamidase: Process Parameters and Removal of Acrylamide from Coffee. Food Chem. 2019, 275, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Im, J.K.; Cho, Y.C.; Noh, H.R.; Yu, S.J. Geographical Distribution and Risk Assessment of Volatile Organic Compounds in Tributaries of the Han River Watershed. Agronomy 2021, 11, 956. [Google Scholar] [CrossRef]
- Hermansson, E.; Schuster, E.; Lindgren, L.; Altskär, A.; Ström, A. Impact of Solvent Quality on the Network Strength and Structure of Alginate Gels. Carbohydr. Polym. 2016, 144, 289–296. [Google Scholar] [CrossRef] [Green Version]
- Baker, J.P.; Bernard, D.P.; Christensen, S.W.; Sale, M.J.; Freda, J.; Heltcher, K.; Rowe, L.; Scanion, P.; Stokes, P.; Suter, G.; et al. Biological Effects of Changes in Surface Water Acid-Base Chemistry; U.S. Department of Energy: Washington, DC, USA, 1990. [CrossRef]
- Li, F.Y.; Xing, Y.J.; Ding, X. Immobilization of Papain on Cotton Fabric by Sol-Gel Method. Enzyme Microb. Technol. 2007, 40, 1692–1697. [Google Scholar] [CrossRef]
- Náray-Szabó, G. Electrostatic Effects in Proteins. Period. Biol. 1999, 101, 325–331. [Google Scholar] [CrossRef]
- Seffernick, J.L.; Reynolds, E.; Fedorov, A.A.; Fedorov, E.; Almo, S.C.; Sadowsky, M.J.; Wackett, L.P. X-Ray Structure and Mutational Analysis of the Atrazine Chlorohydrolase TrzN. J. Biol. Chem. 2010, 285, 30606–30614. [Google Scholar] [CrossRef] [Green Version]
- DeGroot, A.R.; Neufeld, R.J. Encapsulation of Urease in Alginate Beads and Protection from α-Chymotrypsin with Chitosan Membranes. Enzyme Microb. Technol. 2001, 29, 321–327. [Google Scholar] [CrossRef]








| Material | Activity Trypsin Digestion | Activity after Six Week Storage | Thermostability at 60 °C | MeOH as a Co-Solvent (20:80) | Activity at pH 4.0 | Activity at pH 9.0 |
|---|---|---|---|---|---|---|
| Alginate | 75% | 0% | 52% | 10% | 0% | 0% |
| Chitosan | 80% | 20% | 30% | 33% | 0% | 18% |
| Sol-Gel | 85% | 65% | 24% | 24% | 45% | 94% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diviesti, K.; Holz, R.C. Catalytic Biomaterials for Atrazine Degradation. Catalysts 2023, 13, 140. https://doi.org/10.3390/catal13010140
Diviesti K, Holz RC. Catalytic Biomaterials for Atrazine Degradation. Catalysts. 2023; 13(1):140. https://doi.org/10.3390/catal13010140
Chicago/Turabian StyleDiviesti, Karla, and Richard C. Holz. 2023. "Catalytic Biomaterials for Atrazine Degradation" Catalysts 13, no. 1: 140. https://doi.org/10.3390/catal13010140
APA StyleDiviesti, K., & Holz, R. C. (2023). Catalytic Biomaterials for Atrazine Degradation. Catalysts, 13(1), 140. https://doi.org/10.3390/catal13010140