Insights into Solution Structures of Photosynthetic Protein Complexes from Small-Angle Scattering Methods
Abstract
:1. Introduction
2. Basic SANS/SAXS Theory
2.1. SANS/SAXS DATA of Photosynthetic Protein Complexes
2.1.1. PC as Example for a Water-Soluble Protein Studied by SAXS
2.1.2. PSI as Example for a Membrane Protein Studied by SAXS
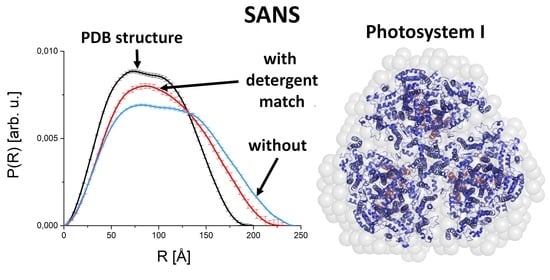
2.1.3. PSI as Example for a Membrane Protein Studied by SANS
3. Solution Structure of the PSI–βDM Complex and Its Detergent Belt
4. Comparison of Different PSI Preparations
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nelson, N.; Yocum, C.F. Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 2006, 57, 521–565. [Google Scholar] [CrossRef] [Green Version]
- Müh, F.; Zouni, A. Light-induced water oxidation in Photosystem II. Front. Biosci. 2011, 16, 3072–3132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambreva, M.D.; Russo, D.; Polticelli, F.; Scognamiglio, V.; Antonacci, A.; Zobnina, V.; Campi, G.; Rea, G. Struc-ture/function/dynamics of Photosystem II plastoquinone binding sites. Curr. Protein Pept. Sci. 2014, 15, 285–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Najafpour, M.M.; Renger, G.; Hołyńska, M.; Moghaddam, A.N.; Aro, E.-M.; Carpentier, R.; Nishihara, H.; Eaton-Rye, J.J.; Shen, J.-R.; Allakhverdiev, S.I. Manganese Compounds as Water-Oxidizing Catalysts: From the Natural Water-Oxidizing Complex to Nanosized Manganese Oxide Structures. Chem. Rev. 2016, 116, 2886–2936. [Google Scholar] [CrossRef]
- Golbeck, J.H. Photosystem I, the Light-Driven Plastocyanin: Ferredoxin Oxidoreductase, 1st ed.; Springer: Dordrecht, The Netherlands, 2006; Volume 24, p. 713. [Google Scholar]
- Mershin, A.; Matsumoto, K.; Kaiser, L.; Yu, D.; Vaughn, M.; Nazeeruddin, K.; Bruce, B.D.; Graetzel, M.; Zhang, S. Self-assembled photosystem-I biophotovoltaics on nanostructured TiO2 and ZnO. Sci. Rep. 2012, 2, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorka, M.; Schartner, J.; van der Est, A.; Rogner, M.; Golbeck, J.H. Light-mediated hydrogen generation in Photosystem I: Attachment of a naphthoquinone-molecular wire-Pt nanoparticle to the A1A and A1B sites. Biochemistry 2014, 53, 2295–2306. [Google Scholar] [CrossRef] [PubMed]
- Feifel, S.C.; Lokstein, H.; Hejazi, M.; Zouni, A.; Lisdat, F. Unidirectional photocurrent of Photosystem I on pi-system-modified graphene electrodes: Nanobionic approaches for the construction of photobiohybrid systems. Langmuir 2015, 31, 10590–10598. [Google Scholar] [CrossRef] [PubMed]
- Stieger, K.R.; Feifel, S.C.; Lokstein, H.; Hejazi, M.; Zouni, A.; Lisdat, F. Biohybrid architectures for efficient light-to-current conversion based on photosystem I within scalable 3D mesoporous electrodes. J. Mater. Chem. A 2016, 4, 17009–17017. [Google Scholar] [CrossRef]
- Jordan, P.; Fromme, P.; Witt, H.T.; Klukas, O.; Saenger, W.; Krauss, N. Three-dimensional structure of cyanobacterial Photo-system I at 2.5 A resolution. Nature 2001, 411, 909–917. [Google Scholar] [CrossRef]
- Mazor, Y.; Borovikova, A.; Caspy, I.; Nelson, N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat. Plants 2017, 3, 17014. [Google Scholar] [CrossRef] [Green Version]
- Gisriel, C.; Coe, J.; Letrun, R.; Yefanov, O.M.; Luna-Chavez, C.; Stander, N.E.; Lisova, S.; Mariani, V.; Kuhn, M.; Aplin, S.; et al. Membrane protein megahertz crystallography at the European XFEL. Nat. Commun. 2019, 10, 5021. [Google Scholar] [CrossRef] [Green Version]
- Young, I.D.; Ibrahim, M.; Chatterjee, R.; Gul, S.; Fuller, F.D.; Koroidov, S.; Brewster, A.S.; Tran, R.; Alonso-Mori, R.; Kroll, T.; et al. Structure of photosystem II and substrate binding at room temperature. Nat. Cell Biol. 2016, 540, 453–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kern, J.; Chatterjee, R.; Young, I.D.; Fuller, F.D.; Lassalle, L.; Ibrahim, M.; Gul, S.; Fransson, T.; Brewster, A.S.; Alonso-Mori, R.; et al. Structures of the intermediates of Kok’s photosynthetic water oxidation clock. Nat. Cell Biol. 2018, 563, 421–425. [Google Scholar] [CrossRef]
- Kölsch, A.; Radon, C.; Golub, M.; Baumert, A.; Bürger, J.; Mielke, T.; Lisdat, F.; Feoktystov, A.; Pieper, J.; Zouni, A.; et al. Current limits of structural biology: The transient interaction between cytochrome c and photosystem I. Curr. Res. Struct. Biol. 2020, 2, 171–179. [Google Scholar] [CrossRef]
- Le Maire, M.; Champeil, P.; Møller, J.V. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta (BBA) Biomembr. 2000, 1508, 86–111. [Google Scholar] [CrossRef] [Green Version]
- Seddon, A.M.; Curnow, P.; Booth, P.J. Membrane proteins, lipids and detergents: Not just a soap opera. Biochim. Biophys. Acta (BBA) Biomembr. 2004, 1666, 105–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mo, Y.; Lee, B.K.; Ankner, J.F.; Becker, J.M.; Heller, W.T. Detergent-associated solution conformations of helical and beta-barrel membrane proteins. J. Phys. Chem. B 2008, 112, 13349–13354. [Google Scholar] [CrossRef] [Green Version]
- Cardoso, M.B.; Smolensky, D.; Heller, W.T.; O’Neill, H. Insight into the Structure of Light-Harvesting Complex II and Its Stabilization in Detergent Solution. J. Phys. Chem. B 2009, 113, 16377–16383. [Google Scholar] [CrossRef]
- Jacques, D.A.; Trewhella, J. Small-angle scattering for structural biology-Expanding the frontier while avoiding the pitfalls. Protein Sci. 2010, 19, 642–657. [Google Scholar] [CrossRef] [Green Version]
- Breyton, C.; Gabel, F.; Lethier, M.; Flayhan, A.; Durand, G.; Jault, J.-M.; Juillan-Binard, C.; Imbert, L.; Moulin, M.; Ravaud, S.; et al. Small angle neutron scattering for the study of solubilised membrane proteins. Eur. Phys. J. E 2013, 36, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kikhney, A.G.; Svergun, D.I. A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 2015, 589, 2570–2577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, P.; Pieper, J.; Kaminskaya, O.; Eckert, H.-J.; Lechner, R.E.; Shuvalov, V.; Renger, G. Reaction pattern of Photosystem II: Oxidative water cleavage and protein flexibility. Photosynth. Res. 2005, 84, 317–323. [Google Scholar] [CrossRef]
- Pieper, J.; Renger, G. Protein dynamics investigated by neutron scattering. Photosynth. Res. 2009, 102, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Pieper, J.; Trapp, M.; Skomorokhov, A.; Natkaniec, I.; Peters, J.; Renger, G. Temperature-dependent vibrational and confor-mational dynamics of Photosystem II membrane fragments from spinach investigated by elastic and inelastic neutron scattering. Biochim. Biophys. Acta 2012, 1817, 1213–1219. [Google Scholar] [CrossRef] [Green Version]
- Golub, M.; Rusevich, L.; Irrgang, K.-D.; Pieper, J. Rigid versus Flexible Protein Matrix: Light-Harvesting Complex II Exhibits a Temperature-Dependent Phonon Spectral Density. J. Phys. Chem. B 2018, 122, 7111–7121. [Google Scholar] [CrossRef]
- Nagy, G.; Garab, G.; Pieper, J. Neutron scattering in photosynthesis research. In Contemporary Problems of Photosynthesis; Al-lakhverdiev, S., Rubin, A.B., Shuvalov, V.A., Eds.; Institute of Computer Science: Izhevsk, Russia, 2014; Volume 1, pp. 69–121. [Google Scholar]
- Tiede, D.M.; Thiyagarjan, P. Characterization of photosynthetic supramolecular assemblies using small angle neutron scat-tering. In Biophysical Techniques in Photosynthesis; Amesz, J., Hoff, A.J., Eds.; Springer: Dordrecht, The Netherlands, 1996; Volume 3, pp. 375–390. [Google Scholar]
- Kirkensgaard, J.J.K.; Holm, J.K.; Larsen, J.K.; Posselt, D. Simulation of small-angle X-ray scattering from thylakoid membranes. J. Appl. Cryst. 2009, 42, 649–659. [Google Scholar] [CrossRef]
- Nagy, G.; Posselt, D.; Kovács, L.; Holm, J.K.; Szabó, M.; Ughy, B.; Rosta, L.; Peters, J.; Timmins, P.; Garab, G. Reversible membrane reorganizations during photosynthesis in vivo: Revealed by small-angle neutron scattering. Biochem. J. 2011, 436, 225–230. [Google Scholar] [CrossRef] [Green Version]
- Liberton, M.; Page, L.E.; O’Dell, W.B.; O’Neill, H.; Mamontov, E.; Urban, V.S.; Pakrasi, H.B. Organization and Flexibility of Cyanobacterial Thylakoid Membranes Examined by Neutron Scattering. J. Biol. Chem. 2013, 288, 3632–3640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakubauskas, D.; Kowalewska, Ł.; Sokolova, A.V.; Garvey, C.J.; Mortensen, K.; Jensen, P.E.; Kirkensgaard, J.J.K. Ultrastructural modeling of small angle scattering from photosynthetic membranes. Sci. Rep. 2019, 9, 19405. [Google Scholar] [CrossRef] [PubMed]
- Tiede, D.M.; Littrell, K.; Marone, P.A.; Zhang, R.; Thiyagarjan, P. Solution structure of a biological bimolecular electron transfer complex: Characterization of the photosynthetic reaction center-cytochrome C2 protein complex by small angle neutron scat-tering. J. Appl. Crystallogr. 2000, 33, 560–564. [Google Scholar] [CrossRef]
- O’Neill, H.; Heller, W.T.; Helton, K.E.; Urban, V.S.; Greenbaum, E. Small-Angle X-ray Scattering Study of Photosystem I−Detergent Complexes: Implications for Membrane Protein Crystallization. J. Phys. Chem. B 2007, 111, 4211–4219. [Google Scholar] [CrossRef] [PubMed]
- Slowik, D.; Rossmann, M.; Konarev, P.V.; Irrgang, K.D.; Saenger, W. Structural investigation of PsbO from plant and cyano-bacterial Photosystem II. J. Mol. Biol. 2011, 407, 125–137. [Google Scholar] [CrossRef]
- Tang, K.-H.; Blankenship, R.E. Neutron and light scattering studies of light-harvesting photosynthetic antenna complexes. Photosynth. Res. 2011, 111, 205–217. [Google Scholar] [CrossRef]
- Le, R.K.; Harris, B.J.; Iwuchukwu, I.J.; Bruce, B.D.; Cheng, X.; Qian, S.; Heller, W.T.; O’Neill, H.; Frymier, P.D. Analysis of the solution structure of Thermosynechococcus elongatus Photosystem I in n-dodecyl-beta-D-maltoside using small-angle neutron scattering and molecular dynamics simulation. Arch. Biochem. Biophys. 2014, 550, 50–57. [Google Scholar] [CrossRef]
- Golub, M.; Moldenhauer, M.; Schmitt, F.-J.; Feoktystov, A.; Mändar, H.; Maksimov, E.; Friedrich, T.; Pieper, J. Solution Structure and Conformational Flexibility in the Active State of the Orange Carotenoid Protein: Part I. Small-Angle Scattering. J. Phys. Chem. B 2019, 123, 9525–9535. [Google Scholar] [CrossRef] [PubMed]
- Golub, M.; Moldenhauer, M.; Schmitt, F.J.; Lohstroh, W.; Maksimov, E.G.; Friedrich, T.; Pieper, J. Solution structure and con-formational flexibility in the active state of the Orange Carotenoid Protein. Part II: Quasielastic neutron scattering. J. Phys. Chem. B 2019, 123, 9536–9545. [Google Scholar] [CrossRef] [PubMed]
- Guinier, A.; Fournet, G. Small-Angle Scattering of X-rays; John Wiley and Sons: New York, NY, USA, 1955. [Google Scholar]
- Svergun, D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Cryst. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Konarev, P.V.; Volkov, V.V.; Sokolova, A.V.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Cryst. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Franke, D.; Svergun, D.I. DAMMIF, a program for rapid ab-Initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009, 42, 342–346. [Google Scholar] [CrossRef] [Green Version]
- Petoukhov, M.V.; Franke, D.; Shkumatov, A.V.; Tria, G.; Kikhney, A.G.; Gajda, M.; Gorba, C.; Mertens, H.D.; Konarev, P.V.; Svergun, D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012, 45, 342–350. [Google Scholar] [CrossRef] [Green Version]
- Svergun, D.I.; Barberato, C.; Koch, M.H.J. CRYSOL—A program to evaluate X-ray solution scattering of biological macro-molecules form atomic coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. [Google Scholar] [CrossRef]
- Svergun, D.I.; Richard, S.; Koch, M.H.J.; Sayers, Z.; Kuprin, S.; Zaccai, G. Protein hydration in solution: Experimental obser-vation by x-ray and neutron scattering. Proc. Natl. Acad. Sci. USA 1998, 95, 2267–2272. [Google Scholar] [CrossRef] [Green Version]
- DeLano, W.L. PyMOL Molecular Graphics System 0.99; Schrödinger, Inc: New York, NY, USA, 2006. [Google Scholar]
- Jacrot, B. The study of biological structures by neutrons cattering from solution. Rep. Prog. Phys. 1976, 39, 911–953. [Google Scholar] [CrossRef]
- Kikhney, A.G.; Borges, C.R.; Molodenskiy, D.S.; Jeffries, C.M.; Svergun, D.I. SASBDB: Towards an automatically curated and validated repository for biological scattering data. Protein Sci. 2020, 29, 66–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feoktystov, A.V.; Frielinghaus, H.; Di, Z.; Jaksch, S.; Pipich, V.; Appavou, M.-S.; Babcock, E.; Hanslik, R.; Engels, R.; Kemmerling, G.; et al. KWS-1 high-resolution small-angle neutron scattering instrument at JCNS: Current state. J. Appl. Cryst. 2015, 48, 61–70. [Google Scholar] [CrossRef]
- Frielinghaus, H.; Feoktystov, A.; Berts, I.; Mangiapia, G. KWS-1: Small-angle scattering diractometer. J. Large Scale Res. Facil. 2015, 1, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Kölsch, A.; Hejazi, M.; Stieger, K.R.; Feifel, S.C.; Kern, J.F.; Müh, F.; Lisdat, F.; Lokstein, H.; Zouni, A. Insights into the binding behavior of native and non-native cytochromes to photosystem I from Thermosynechococcus elongatus. J. Biol. Chem. 2018, 293, 9090–9100. [Google Scholar] [CrossRef] [Green Version]
- Kern, J.; Loll, B.; Luneberg, C.; DiFiore, D.; Biesiadka, J.; Irrgang, K.D.; Zouni, A. Purification, characterisation and crystalli-sation of Photosystem II from Thermosynechococcus elongatus cultivated in a new type of photobioreactor. Biochim. Biophys. Acta 2005, 1706, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Fromme, R.; Ishchenko, A.; Metz, M.; Chowdhury, S.R.; Basu, S.; Boutet, S.; Fromme, P.; White, T.A.; Barty, A.; Spence, J.C.H.; et al. Serial femtosecond crystallography of soluble proteins in lipidic cubic phase. IUCrJ 2015, 2, 545–551. [Google Scholar] [CrossRef] [Green Version]
- Conrad, C.E.; Basu, S.E.; James, D.; Wang, D.; Schaffer, A.; Roy-Chowdhury, S.; Zatsepin, N.A.; Aquila, A.; Coe, J.; Gati, C.; et al. A novel inert crystal delivery medium for serial femtosecond crystallography. IUCrJ 2015, 2, 421–430. [Google Scholar] [CrossRef]
- Chen, M.; Floetenmeyer, M.; Bibby, T.S. Supramolecular organization of phycobiliproteins in the chlorophyll d -containing cyanobacterium Acaryochloris marina. FEBS Lett. 2009, 583, 2535–2539. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Ikeuchi, M. Phycobilisome: Architecture of a light-harvesting supercomplex. Photosynth. Res. 2013, 116, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Theiss, C.; Schmitt, F.-J.; Pieper, J.; Nganou, C.; Grehn, M.; Vitali, M.; Olliges, R.; Eichler, H.J.; Eckert, H.-J. Excitation energy transfer in intact cells and in the phycobiliprotein antennae of the chlorophyll d containing cyanobacterium Acaryochloris marina. J. Plant Physiol. 2011, 168, 1473–1487. [Google Scholar] [CrossRef]
- Gryliuk, G.; Rätsep, M.; Hildebrandt, S.; Irrgang, K.-D.; Eckert, H.-J.; Pieper, J. Excitation energy transfer and electron-vibrational coupling in phycobiliproteins of the cyanobacterium Acaryochloris marina investigated by site-selective spectroscopy. Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1837, 1490–1499. [Google Scholar] [CrossRef] [Green Version]
- Pieper, J.; Rätsep, M.; Golub, M.; Schmitt, F.-J.; Artene, P.; Eckert, H.-J. Excitation energy transfer in phycobiliproteins of the cyanobacterium Acaryochloris marina investigated by spectral hole burning. Photosynth. Res. 2017, 133, 225–234. [Google Scholar] [CrossRef]
- Golub, M.; Combet, S.; Wieland, D.; Soloviov, D.; Kuklin, A.; Lokstein, H.; Schmitt, F.-J.; Olliges, R.; Hecht, M.; Eckert, H.-J.; et al. Solution structure and excitation energy transfer in phycobiliproteins of Acaryochloris marina investigated by small angle scattering. Biochim. Biophys. Acta (BBA) Bioenerg. 2017, 1858, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Golub, M.; Hejazi, M.; Kölsch, A.; Lokstein, H.; Wieland, D.C.F.; Zouni, A.; Pieper, J. Solution structure of monomeric and trimeric photosystem I of Thermosynechococcus elongatus investigated by small-angle X-ray scattering. Photosynth. Res. 2017, 133, 163–173. [Google Scholar] [CrossRef] [Green Version]
- Golub, M.; Hussein, R.; Ibrahim, M.; Hecht, M.; Wieland, D.C.F.; Martel, A.; Machado, B.; Zouni, A.; Pieper, J. Solution Structure of the Detergent–Photosystem II Core Complex Investigated by Small-Angle Scattering Techniques. J. Phys. Chem. B 2020, 124, 8583–8592. [Google Scholar] [CrossRef] [PubMed]
- Vasilev, S.; Irrgang, K.D.; Schrotter, T.; Bergmann, A.; Eichler, H.J.; Renger, G. Quenching of chlorophyll alpha fluorescence in the aggregates of LHCII: Steady state fluorescence and picosecond relaxation kinetics. Biochemistry 1997, 36, 7503–7512. [Google Scholar] [CrossRef]
- Pieper, J.; Irrgang, K.-D.; Rätsep, M.; Jankowiak, R.; Schrötter, T.; Voigt, J.; Small, G.J.; Renger, G. Effects of Aggregation on Trimeric Light-Harvesting Complex II of Green Plants: A Hole-Burning Study. J. Phys. Chem. A 1999, 103, 2422–2428. [Google Scholar] [CrossRef]
- Voigt, B.; Krikunova, M.; Lokstein, H. Influence of detergent concentration on aggregation and spectroscopic properties of light-harvesting complex II. Photosynth. Res. 2007, 95, 317–325. [Google Scholar] [CrossRef]
- Midtgaard, S.R.; Darwish, T.A.; Pedersen, M.C.; Huda, P.; Larsen, A.H.; Jensen, G.V.; Kynde, S.A.R.; Skar-Gislinge, N.; Nielsen, A.J.Z.; Olesen, C.; et al. Invisible detergents for structure determination of membrane proteins by small-angle neutron scattering. FEBS J. 2017, 285, 357–371. [Google Scholar] [CrossRef]
- Juers, D.H.; Matthews, B.W. Reversible lattice repacking illustrates the temperature dependence of macromolecular interac-tions. J. Mol. Biol. 2001, 311, 851–862. [Google Scholar] [CrossRef] [Green Version]
- Malferrari, M.; Savitsky, A.; Mamedov, M.D.; Milanovsky, G.E.; Lubitz, W.; Möbius, K.; Semenov, A.Y.; Venturoli, G. Trehalose matrix effects on charge-recombination kinetics in Photosystem I of oxygenic photosynthesis at different dehydration levels. Biochim. Biophys. Acta (BBA) Bioenerg. 2016, 1857, 1440–1454. [Google Scholar] [CrossRef]
- Pieper, J.; Hauß, T.; Buchsteiner, A.; Renger, G. The effect of hydration on protein flexibility in photosystem II of green plants studied by quasielastic neutron scattering. Eur. Biophys. J. 2008, 37, 657–663. [Google Scholar] [CrossRef]
- Russo, D.; Lambreva, M.D.; Simionesco, C.A.; Sebban, P.; Rea, G. Dynamics Properties of Photosynthetic Microorganisms Probed by Incoherent Neutron Scattering. Biophys. J. 2019, 116, 1759–1768. [Google Scholar] [CrossRef]
- Møller, J.; Le Maire, M. Detergent binding as a measure of hydrophobic surface area of integral membrane proteins. J. Biol. Chem. 1993, 268, 18659–18672. [Google Scholar] [CrossRef]
- Pebay-Peyroula, E.; Garavito, R.M.; Rosenbusch, J.P.; Zulauf, M.A.; Timmins, P. Detergent structure in tetragonal crystals of OmpF porin. Structure 1995, 3, 1051–1059. [Google Scholar] [CrossRef] [Green Version]
- Müh, F.; Zouni, A. Micelle formation in the presence of photosystem I. Biochim. Biophys. Acta (BBA) Biomembr. 2008, 1778, 2298–2307. [Google Scholar] [CrossRef] [Green Version]
- Hussein, R.; Ibrahim, M.; Chatterjee, R.; Coates, L.; Müh, F.; Yachandra, V.K.; Yano, J.; Kern, J.; Dobbek, H.; Zouni, A. Opti-mizing crystal size of Photosystem II by macroseeding: Toward neutron protein crystallography. Cryst. Growth Des. 2018, 18, 85–94. [Google Scholar] [CrossRef] [PubMed]







| SANS | SAXS | |
|---|---|---|
| Incident beam source | Neutrons | Photons (X-ray) |
| Interacting field | Nuclei | Electrons |
| Incident beam wavelength (Å) | 4–25 | 1–1.5 |
| Incident beam flux | 108–109 neutrons/cm2/s | 1011 photons/cm2/s |
| Typical sample counting time | Minutes to hours | Seconds to minutes |
| Contrast variation | Yes | No |
| Radiation damage | No | High (synchrotron radiation) |
| Sample amount required | High | Medium |
| Availability | Large facility only | Laboratory and synchrotron radiation sources |
| PDB | Reference | Guinier Rg (Å) | Rg (Å) from IFT (Gnom) | Dmax (Å) | |
|---|---|---|---|---|---|
| SANS PSI 5% D2O | Figure 3 | 75.2 ± 3 | 75.8 ± 3 | 230 ± 10 | |
| SANS PSI 100% D2O | Figure 3 and Figure 5 | 79 ± 3 | 83 ± 3 | 245 ± 10 | |
| SAXS PSI monomer | [62] | 58 ± 4 | 54 ± 2 | 185 ± 10 | |
| SAXS PSI trimer | [62] | 78 ± 2 | 79.8 ± 2 | 250 ± 15 | |
| PSI T. elongatus (trimer) crystal structure | 1JB0 | [10] | 68.2 | 68.28 | 200 |
| SANS PSI 18% D2O | [37] | 77.9 ± 2.86 | 75.9 ± 0.1 | 215 ± 10 | |
| SANS PSI 100% D2O | [37] | 94.9 ± 2.32 | 93.1 ± 1.1 | 280 ± 10 | |
| Plant PSI (monomer) crystal structure | 5L8R | [11] | 50.3 | 50.3 | 173.8 |
| Plant PSI d-βDM 100% D2O (monomer) | [67] | 55.2 ± 0.2 | 199 ± 5 | ||
| PSI T. elongatus (trimer) Cryo-EM | 6TRD | [15] | 68.15 | 68.51 | 205 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golub, M.; Kölsch, A.; Feoktystov, A.; Zouni, A.; Pieper, J. Insights into Solution Structures of Photosynthetic Protein Complexes from Small-Angle Scattering Methods. Crystals 2021, 11, 203. https://doi.org/10.3390/cryst11020203
Golub M, Kölsch A, Feoktystov A, Zouni A, Pieper J. Insights into Solution Structures of Photosynthetic Protein Complexes from Small-Angle Scattering Methods. Crystals. 2021; 11(2):203. https://doi.org/10.3390/cryst11020203
Chicago/Turabian StyleGolub, Maksym, Adrian Kölsch, Artem Feoktystov, Athina Zouni, and Jörg Pieper. 2021. "Insights into Solution Structures of Photosynthetic Protein Complexes from Small-Angle Scattering Methods" Crystals 11, no. 2: 203. https://doi.org/10.3390/cryst11020203
APA StyleGolub, M., Kölsch, A., Feoktystov, A., Zouni, A., & Pieper, J. (2021). Insights into Solution Structures of Photosynthetic Protein Complexes from Small-Angle Scattering Methods. Crystals, 11(2), 203. https://doi.org/10.3390/cryst11020203