Sequence Analysis and Preliminary X-ray Crystallographic Analysis of an Acetylesterase (LgEstI) from Lactococcus garvieae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Clustering
2.2. Esterase Activity
2.3. Gene Cloning, Expression, and Purification of Recombinant LgEstI Protein
2.4. LgEstI Crystallization, Data Collection, and Phasing
3. Results
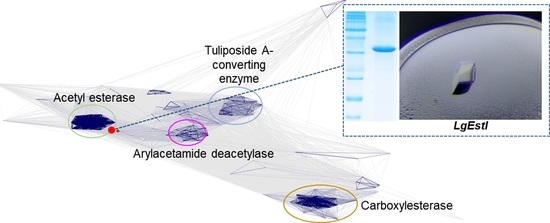
3.1. Clustering Analysis of LgEstI
3.2. Purification and Biochemical Characterization of LgEstI
3.3. X-ray Crystallographic Study of LgEstI
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fazary, A.E.; Ju, Y.H. Feruloyl Esterases as Biotechnological Tools: Current and Future Perspectives. Acta Biochim. Biophys. Sin. 2007, 39, 811–828. [Google Scholar] [CrossRef] [PubMed]
- Celligoi, M.A.P.C.; Baldo, C.; De Melo, M.R.; Gasparin, F.G.M.; Marques, T.A.; De Barros, M. Lipase Properties, Functions and Food Applications, Microb. In Microbial Enzyme Technology in Food Applications; CRC Press: Boca Raton, FL, USA, 2017; pp. 214–240. [Google Scholar] [CrossRef]
- Pleiss, J.; Fischer, M.; Peiker, M.; Thiele, C.; Schmid, R.D. Lipase Engineering Database. J. Mol. Catal. B Enzym. 2000, 10, 491–508. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. Microbial carboxyl esterases: Classification, properties and application in biocatalysis. FEMS Microbiol. Rev. 2002, 26, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Barzkar, N.; Sohail, M.; Tamadoni Jahromi, S.; Gozari, M.; Poormozaffar, S.; Nahavandi, R.; Hafezieh, M. Marine Bacterial Esterases: Emerging Biocatalysts for Industrial Applications. Appl. Biochem. Biotechnol. 2021, 193, 1187–1214. [Google Scholar] [CrossRef] [PubMed]
- Larsen, E.M.; Johnson, R.J. Microbial esterases and ester prodrugs: An unlikely marriage for combating antibiotic resistance. Drug Dev. Res. 2019, 80, 33–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, S.; Yoo, W.; Kim, Y.O.; Kim, K.K.; Kim, T.D. Molecular Characterization of a Novel Family VIII Esterase with β-Lactamase Activity (PsEstA) from Paenibacillus sp. Biomolecules 2019, 9, 786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morita, H.; Toh, H.; Oshima, K.; Yoshizaki, M.; Kawanishi, M.; Nakaya, K.; Suzuki, T.; Miyauchi, E.; Ishii, Y.; Tanabe, S.; et al. Complete genome sequence and comparative analysis of the fish pathogen Lactococcus garvieae. PLoS ONE 2011, 6, e23184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabler, F.; Nam, S.Z.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinform. 2020, 72, e108. [Google Scholar] [CrossRef] [PubMed]
- Frickey, T.; Lupas, A. CLANS: A Java Application for Visualizing Protein Families Based on Pairwise Similarity. Bioinformatics 2004, 20, 3702–3704. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Kwon, S.; Park, S.H.; Kim, B.Y.; Yoo, W.; Ryu, B.H.; Kim, H.W.; Shin, S.C.; Kim, S.; Park, H.; et al. Crystal Structure and Functional Characterization of an Esterase (EaEST) from Exiguobacterium antarcticum. PLoS ONE 2017, 12, e0169540. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karplus, P.A.; Diederichs, K. Linking Crystallographic Model and Data Quality. Science 2012, 336, 1030–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| LgEst | |
|---|---|
| Source organism | Lactococcus garvieae |
| DNA source | Genomic DNA |
| Cloning vector | pET21a |
| Expression host | Escherichia coli BL21 (DE3) |
| Amino acid sequence | MVERISLEKAALEFSEANAPHPRIYELPVEEGRSLLNEV QDSPVVKEDVDIEDIAVDTGEWGEINVRFIRPLHQEKKL PVIFYIHGAGWVFGNAHTHDKLIRELAVRTNSVVVFSE YSLSPEAKYPTAIEQNYAVLQQLKDFANDKKFDVNHLT VAGDSVGGNMATVMTLLTKQRGGQKIGQQVLYYPVT DANFDTDSYNEFAENYFLTKEGMIWFWDQYTTSQEER HQITASPLRATKEDLADLPAALIITGEADVLRDEGEAYA RKLREADVEVTQVRFQAIIHDFVMVNSMNETHATRAA MSLSTQWINEKNRK |
| Method | Vapor Diffusion |
|---|---|
| Plate type for screening | 96-well sitting drop MRC plate (Molecular Dimension, Suffolk, UK) |
| Plate type for optimization | 24-well hanging drop plate (Molecular Dimension, Suffolk, UK) |
| Temperature (°C) | 22 |
| Protein concentration (mg/mL) | 25 |
| Composition of protein solution | 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 1 mM TECP |
| Composition of reservoir solution | 0.1 M Tris: HCl (pH 7.1), 0.2 M calcium acetate hydrate, and 19% (w/v) PEG 3000 |
| Volume and ratio of drop | 2 μL, 1:1 |
| Volume of reservoir (μL) | 500 |
| Data Collection | LgEstI |
|---|---|
| Wavelength (Å) | 0.97949 |
| X-ray source | BL-5C beamline |
| Rotation range per image (°) | 1 |
| Exposure time (s) | 0.1 |
| Space group | I21 |
| Unit-cell parameters (Å, °) | a = 54.41, b = 92.77, c = 218.11, α = γ = 90.0, β = 96.61 |
| Resolution range (Å) a | 28.49–2.0 (2.05–2.0) |
| No. of observed reflections a | 494,404 |
| No. of unique reflections a | 71,847 (4652) |
| Completeness (%) a | 98.7 (99.8) |
| Redundancy a | 6.9 (7.2) |
| Rsyma,b | 0.059 (0.540) |
| I/σ a | 16.1 (2.8) |
| Mosaicity | 0.36 |
| CC½ a,c | (99.7%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Do, H.; Wang, Y.; Lee, C.W.; Yoo, W.; Jeon, S.; Hwang, J.; Lee, M.J.; Kim, K.K.; Kim, H.-W.; Lee, J.H.; et al. Sequence Analysis and Preliminary X-ray Crystallographic Analysis of an Acetylesterase (LgEstI) from Lactococcus garvieae. Crystals 2022, 12, 46. https://doi.org/10.3390/cryst12010046
Do H, Wang Y, Lee CW, Yoo W, Jeon S, Hwang J, Lee MJ, Kim KK, Kim H-W, Lee JH, et al. Sequence Analysis and Preliminary X-ray Crystallographic Analysis of an Acetylesterase (LgEstI) from Lactococcus garvieae. Crystals. 2022; 12(1):46. https://doi.org/10.3390/cryst12010046
Chicago/Turabian StyleDo, Hackwon, Ying Wang, Chang Woo Lee, Wanki Yoo, Sangeun Jeon, Jisub Hwang, Min Ju Lee, Kyeong Kyu Kim, Han-Woo Kim, Jun Hyuck Lee, and et al. 2022. "Sequence Analysis and Preliminary X-ray Crystallographic Analysis of an Acetylesterase (LgEstI) from Lactococcus garvieae" Crystals 12, no. 1: 46. https://doi.org/10.3390/cryst12010046
APA StyleDo, H., Wang, Y., Lee, C. W., Yoo, W., Jeon, S., Hwang, J., Lee, M. J., Kim, K. K., Kim, H. -W., Lee, J. H., & Kim, T. D. (2022). Sequence Analysis and Preliminary X-ray Crystallographic Analysis of an Acetylesterase (LgEstI) from Lactococcus garvieae. Crystals, 12(1), 46. https://doi.org/10.3390/cryst12010046