Synthesis of Molecularly Imprinted Cryogels to Deplete Abundant Proteins from Bovine Serum
Abstract
:1. Introduction
2. Experimental Section
2.1. Instruments and Reagents
2.2. Preparation of Modified Polyacrylamide Cryogels
2.3. Conductometric Titrations of the Amphoteric Cryogels
2.4. Protein Adsorption on the Cryogels
2.5. High Performance Liquid Chromatography (HPLC)
2.6. SDS Polyacrylamide Gel Electrophoresis (SDS-PAGE)
3. Results and Discussion
3.1. Morphological, Structural and Physicochemical Characterization of Cryogels
3.2. Influencing Factors on Adsorption and Imprinting
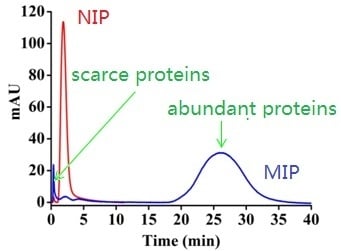
3.3. Chromatography and Electrophoresis Results
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wulff, G.; Gross, T.; Schonfeld, R. Enzyme models based on molecularly imprinted polymers with strong esterase activity. Angew. Chem. Int. Ed. Engl. 1997, 36, 1962–1964. [Google Scholar] [CrossRef]
- Wulff, G. Enzyme-like catalysis by molecularly imprinted polymers. Chem. Rev. 2002, 102, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Ma, Y.; Pan, J.M.; Meng, Z.H.; Pan, G.Q.; Sellergren, B. Molecularly imprinted polymers with stimuli-responsive affinity: Progress and perspectives. Polymers 2015, 7, 1689–1715. [Google Scholar] [CrossRef]
- Wackerlig, J.; Schirhagl, R. Applications of molecularly imprinted polymer nanoparticles and their advances toward industrial use: A review. Anal. Chem. 2016, 88, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; Karimi, M. Novel developments and trends of analytical methods for drug analysis in biological and environmental samples by molecularly imprinted polymers. Trends Anal. Chem. 2017, 89, 146–162. [Google Scholar] [CrossRef]
- Ashley, J.; Shahbazi, M.A.; Kant, K.; Chidambara, V.A.; Wolff, A.; Bang, D.D.; Sun, Y. Molecularly imprinted polymers for sample preparation and biosensing in food analysis: Progress and perspectives. Biosens. Bioelectron. 2017, 91, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, Y.H.; Jiang, Y.D.; Li, S.; Liu, W. Molecularly imprinted polymers for the identification and separation of chiral drugs and biomolecules. Polymers 2016, 8, 216. [Google Scholar] [CrossRef]
- Asman, S.; Mohamad, S.; Sarih, N.M. Effects of raft agent on the selective approach of molecularly imprinted polymers. Polymers 2015, 7, 484–503. [Google Scholar] [CrossRef]
- Asliyuce, S.; Uzun, L.; Rad, A.Y.; Unal, S.; Say, R.; Denizli, A. Molecular imprinting based composite cryogel membranes for purification of anti-hepatitis b surface antibody by fast protein liquid chromatography. J. Chromatogr. B 2012, 889, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, L.; Levine, M. Biomimetic catalysis. Acs Catal. 2011, 1, 1090–1118. [Google Scholar] [CrossRef]
- Ahmad, R.; Griffete, N.; Lamouri, A.; Felidj, N.; Chehimi, M.M.; Mangeney, C. Nanocomposites of gold nanoparticles@molecularly imprinted polymers: Chemistry, processing, and applications in sensors. Chem. Mater. 2015, 27, 5464–5478. [Google Scholar] [CrossRef]
- Yang, K.G.; Zhang, L.H.; Liang, Z.; Zhang, Y.K. Protein-imprinted materials: Rational design, application and challenges. Anal. Bioanal. Chem. 2012, 403, 2173–2183. [Google Scholar] [CrossRef] [PubMed]
- Wulff, G.; Liu, J.Q. Design of biomimetic catalysts by molecular imprinting in synthetic polymers: The role of transition state stabilization. Acc. Chem. Res. 2012, 45, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Culver, H.R.; Peppas, N.A. Protein-imprinted polymers: The shape of things to come? Chem. Mater. 2017, 29, 5753–5761. [Google Scholar] [CrossRef]
- Lv, Y.Q.; Tan, T.W.; Svec, F. Molecular imprinting of proteins in polymers attached to the surface of nanomaterials for selective recognition of biomacromolecules. Biotechnol. Adv. 2013, 31, 1172–1186. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Q.M.; Tian, L.L.; Yang, C.; Yu, S.H.; Yang, C. Research progress of the molecularly imprinted cryogel. Chin. J. Anal. Chem. 2015, 43, 1777–1784. [Google Scholar] [CrossRef]
- Andac, M.; Galaev, I.Y.; Denizli, A. Affinity based and molecularly imprinted cryogels: Applications in biomacromolecule purification. J. Chromatogr. B 2016, 1021, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Asliyuce, S.; Uzun, L.; Say, R.; Denizli, A. Immunoglobulin g recognition with f-ab fragments imprinted monolithic cryogels: Evaluation of the effects of metal-ion assisted-coordination of template molecule. React. Funct. Polym. 2013, 73, 813–820. [Google Scholar] [CrossRef]
- Fatoni, A.; Numnuam, A.; Kanatharana, P.; Limbut, W.; Thavarungkul, P. A novel molecularly imprinted chitosan-acrylamide, graphene, ferrocene composite cryogel biosensor used to detect microalbumin. Analyst 2014, 139, 6160–6167. [Google Scholar] [CrossRef] [PubMed]
- Bereli, N.; Andac, M.; Baydemir, G.; Say, R.; Galaev, I.Y.; Denizli, A. Protein recognition via ion-coordinated molecularly imprinted supermacroporous cryogels. J. Chromatogr. A 2008, 1190, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Tamahkar, E.; Bereli, N.; Say, R.; Denizli, A. Molecularly imprinted supermacroporous cryogels for cytochrome c recognition. J. Sep. Sci. 2011, 34, 3433–3440. [Google Scholar] [CrossRef] [PubMed]
- Cimen, D.; Denizli, A. Immobilized metal affinity monolithic cryogels for cytochrome c purification. Colloids Surf. B 2012, 93, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Corman, M.E.; Armutcu, C.; Ozkara, S.; Uzun, L.; Denizli, A. Molecularly imprinted cryogel cartridges for the specific filtration and rapid separation of interferon alpha. RSC Adv. 2015, 5, 45015–45026. [Google Scholar] [CrossRef]
- Erol, K.; Kose, K.; Uzun, L.; Say, R.; Denizli, A. Polyethyleneimine assisted-two-step polymerization to develop surface imprinted cryogels for lysozyme purification. Colloids Surf. B 2016, 146, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Derazshamshir, A.; Baydemir, G.; Andac, M.; Say, R.; Galaev, I.Y.; Denizli, A. Molecularly imprinted phema-based cryogel for depletion of hemoglobin from human blood. Macromol. Chem. Phys. 2010, 211, 657–668. [Google Scholar] [CrossRef]
- Yang, C.; Liu, G.-F.; Zhou, X.-L.; Liu, Y.-R.; Wang, J.; Tian, L.-L.; Hu, X.-Y.; Wang, Y.-Y. Polyacrylamide based cryogels as catalysts for biodiesel. Catal. Lett. 2015, 145, 1778–1783. [Google Scholar] [CrossRef]
- Yang, C.; Liu, Y.-R.; Zhang, Y.; Wang, J.; Tian, L.-L.; Yan, Y.-N.; Cao, W.-Q.; Wang, Y.-Y. Ice squeezing induced multicolor fluorescence emissions from polyacrylamide cryogels. J. Fluoresc. 2017, in press. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhou, X.-L.; Liu, Y.-R.; Zhang, Y.; Wang, J.; Tian, L.-L.; Yan, Y.-N. Extensive imprinting adaptability of polyacrylamide-based amphoteric cryogels against protein molecules. Chin. J. Anal. Chem. 2016, 44, 1322–1327. [Google Scholar] [CrossRef]
- Yang, C.; Zhou, X.-L.; Liu, Y.-R.; Wang, J.; Tian, L.-L.; Zhang, Y.; Hu, X.-Y. Charged groups synergically enhance protein imprinting in amphoteric polyacrylamide cryogels. J. Appl. Polym. Sci. 2016, 133, 43851. [Google Scholar] [CrossRef]
- Yang, C.; Liu, Y.-R.; Zhang, Y.; Wang, J.; Tian, L.-L.; Yan, Y.-N.; Cao, W.-Q.; Wang, Y.-Y. Depletion of abundant human serum proteins by per se imprinted cryogels based on sample heterogeneity. Proteomics 2017, 17, 1600284. [Google Scholar] [CrossRef] [PubMed]
- Karfa, P.; Madhuri, R.; Sharma, P.K. A battle between spherical and cube-shaped ag/agcl nanoparticle modified imprinted polymer to achieve femtogram detection of alpha-feto protein. J. Mater. Chem. B 2016, 4, 5534–5547. [Google Scholar] [CrossRef]
- Yongabi, D.; Khorshid, M.; Losada-Perez, P.; Eersels, K.; Deschaume, O.; D’Haen, J.; Bartic, C.; Hooyberghs, J.; Thoelen, R.; Wubbenhorst, M.; et al. Cell detection by surface imprinted polymers sips: A study to unravel the recognition mechanisms. Sens. Actuators B Chem. 2018, 255, 907–917. [Google Scholar] [CrossRef]
- Zhang, Z.J.; Zhang, X.H.; Liu, B.W.; Liu, J.W. Molecular imprinting on inorganic nanozymes for hundred-fold enzyme specificity. J. Am. Chem. Soc. 2017, 139, 5412–5419. [Google Scholar] [CrossRef] [PubMed]
- Rebelo, M.; Santos, C.; Costa-Rodrigues, J.; Fernandes, M.H.; Noronha, J.P.; Sales, M.G.F. Novel prostate specific antigen plastic antibody designed with charged binding sites for an improved protein binding and its application in a biosensor of potentiometric transduction. Electrochim. Acta 2014, 132, 142–150. [Google Scholar] [CrossRef]









| Reagents | B | C | D | E | F |
|---|---|---|---|---|---|
| Bovine serum/mL | 0 | 1 | 2 | 4 | 10 |
| H2O/mL | 20 | 19 | 18 | 16 | 10 |
| AM/g | 0.5 | ||||
| BisAM/g | 0.3 | ||||
| Acrylic acid/μL | 125 | ||||
| Diallylamine/μL | 63 | ||||
| VTEOS/μL | 150 | ||||
| SHS/g | 0.03 | ||||
| APS/g | 0.06 | ||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Zhang, Y.; Cao, W.-Q.; Ji, X.-F.; Wang, J.; Yan, Y.-N.; Zhong, T.-L.; Wang, Y. Synthesis of Molecularly Imprinted Cryogels to Deplete Abundant Proteins from Bovine Serum. Polymers 2018, 10, 97. https://doi.org/10.3390/polym10010097
Yang C, Zhang Y, Cao W-Q, Ji X-F, Wang J, Yan Y-N, Zhong T-L, Wang Y. Synthesis of Molecularly Imprinted Cryogels to Deplete Abundant Proteins from Bovine Serum. Polymers. 2018; 10(1):97. https://doi.org/10.3390/polym10010097
Chicago/Turabian StyleYang, Chun, Yan Zhang, Wei-Qin Cao, Xiao-Feng Ji, Jian Wang, Ya-Nan Yan, Tao-Lin Zhong, and Yu Wang. 2018. "Synthesis of Molecularly Imprinted Cryogels to Deplete Abundant Proteins from Bovine Serum" Polymers 10, no. 1: 97. https://doi.org/10.3390/polym10010097
APA StyleYang, C., Zhang, Y., Cao, W. -Q., Ji, X. -F., Wang, J., Yan, Y. -N., Zhong, T. -L., & Wang, Y. (2018). Synthesis of Molecularly Imprinted Cryogels to Deplete Abundant Proteins from Bovine Serum. Polymers, 10(1), 97. https://doi.org/10.3390/polym10010097