Novel Exopolysaccharide Produced from Fermented Bamboo Shoot-Isolated Lactobacillus Fermentum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material
2.2. Isolation of Lactic Acid Bacteria
2.3. Screening Isolate Having High Ability of EPS Production
2.4. EPS Production and Purification
2.5. Identification of LAB by Phenylalanyl-tRNA Synthase (pheS) Gene Sequencing
2.5.1. Preparing DNA Extracts
2.5.2. PCR and Sequencing
2.5.3. Sequence Analysis
2.6. Estimation of EPS Molecular Mass
2.7. Monosaccharide Composition and Methylation Analysis
2.7.1. Methylation Analysis
2.7.2. EPS Hydrolysis
2.7.3. Converting
2.7.4. Acetylation
2.7.5. GC-MS
2.8. NMR Method
2.9. Statistical Analysis
3. Results and Discussion
3.1. Ability of EPS Production of Isolates
3.2. Average Molecular Weight
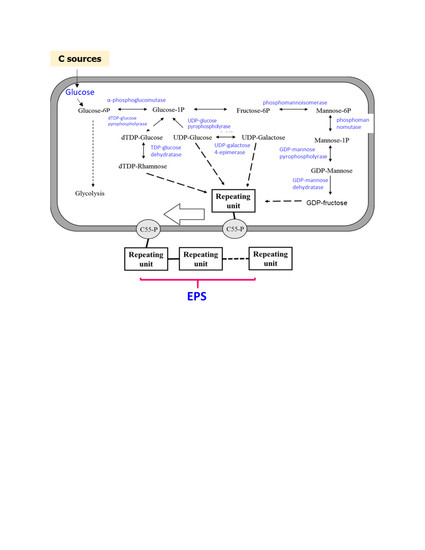
3.3. Methylation Analysis of EPS-MC3
3.4. 1H NMR and 13C NMR Analysis of EPS-MC3
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Harutoshi, T. Exopolysaccharides of lactic acid bacteria for food and colon health applications. Lact. Acid Bact. -R D Food Health Livest. Purp. 2013. [Google Scholar] [CrossRef] [Green Version]
- Patel, A.; Prajapat, J. Food and health applications of exopolysaccharides produced by lactic acid bacteria. Adv. Dairy Res. 2013. [Google Scholar] [CrossRef]
- Fuller, R. Handbook of Probiotics. Int. J. Food Sci. Tech. 2001, 36, 224. [Google Scholar] [CrossRef]
- Parvez, S.; Malik, K.A.; Ah Kang, S.; Kim, H.Y. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 2006, 100, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Saikali, J.; Picard, C.; Freitas, M.; Holt, P. Fermented milks, probiotic cultures, and colon cancer. Nutr. Cancer 2004, 49, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Thirabunyanon, M.; Boonprasom, P.; Niamsup, P. Probiotic potential of lactic acid bacteria isolated from fermented dairy milks on antiproliferation of colon cancer cells. Biotechnol. Lett 2009, 31, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Ismail, B.; Nampoothiri, K.M. Molecular characterization of an exopolysaccharide from a probiotic Lactobacillus plantarum MTCC 9510 and its efficacy to improve the texture of starchy food. J. Food Sci. Technol. 2014, 51, 4012–4018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Zhao, X.; Yang, Y.; Zhao, A.; Yang, Z. Characterization and bioactivities of an exopolysaccharide produced by Lactobacillus plantarum YW32. Int. J. Biol. Macromol. 2015, 74, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, W.; Rui, X.; Chen, X.; Jiang, M.; Dong, M. Characterization of a novel exopolysaccharide with antitumor activity from Lactobacillus plantarum 70810. Int. J. Biol. Macromol. 2014, 63, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Górska-Frączek, S.; Sandström, C.; Kenne, L.; Rybka, J.; Strus, M.; Heczko, P.; Gamian, A. Structural studies of the exopolysaccharide consisting of a nonasaccharide repeating unit isolated from Lactobacillus rhamnosus KL37B. Carbohydr. Res. 2011, 346, 2926–2932. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.K.; Michaud, P.; Singhania, R.R.; Soccol, C.R.; Pandey, A. Polysaccharides from probiotics: New developments as food additives. Food Technol. Biotechnol. 2010, 48, 451–463. [Google Scholar]
- Desiye, A.; Abegaz, K. Isolation, characterization and identification of lactic acid bacteria and yeast involved in fermentation of teff (Eragrostis tef) batter. Adv. Res. Biol. Sci. 2013, 1, 36–44. [Google Scholar]
- Yuliana, N.; Dizon, E.I. Phenotypic identification of lactic acid bacteria isolated from Tempoyak (fermented durian) made in the Philippines. Int. J. Biol. 2011, 3, 145. [Google Scholar] [CrossRef] [Green Version]
- Dec, M.; Urban-Chmiel, R.; Gnat, S.; Puchalski, A.; Wernicki, A. Identification of Lactobacillus strains of goose origin using MALDI-TOF mass spectrometry and 16S–23S rDNA intergenic spacer PCR analysis. Res. Microbiol. 2014, 165, 190–201. [Google Scholar] [CrossRef]
- Liu, A.; Jia, Y.; Zhao, L.; Gao, Y.; Liu, G.; Chen, Y.; Zhao, G.; Xu, L.; Shen, L.; Liu, Y. Diversity of isolated lactic acid bacteria in Ya’an sourdoughs and evaluation of their exopolysaccharide production characteristics. LWT 2018, 95, 17–22. [Google Scholar] [CrossRef]
- Naser, S.M.; Dawyndt, P.; Hoste, B.; Gevers, D.; Vandemeulebroecke, K.; Cleenwerck, I.; Vancanneyt, M.; Swings, J. Identification of Lactobacilli by pheS and rpoA gene sequence analyses. Int. J. Syst. Evol. Microbiol. 2007, 57, 2777–2789. [Google Scholar] [CrossRef] [Green Version]
- Öz, E.; Kaban, G.; Barış, Ö.; Kaya, M. Isolation and identification of lactic acid bacteria from pastırma. Food Control 2017, 77, 158–162. [Google Scholar] [CrossRef]
- Gevers, D.; Huys, G.; Swings, J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. Fems Microbiol. Lett. 2001, 205, 31–36. [Google Scholar] [CrossRef] [Green Version]
- Maldonado, N.C.; Ficoseco, C.A.; Mansilla, F.I.; Melián, C.; Hébert, E.M.; Vignolo, G.M.; Nader-Macías, M.E.F. Identification, characterization and selection of autochthonous lactic acid bacteria as probiotic for feedlot cattle. Livest. Sci. 2018, 212, 99–110. [Google Scholar] [CrossRef]
- Nguyen, H.; Elegado, F.; Librojo-Basilio, N.; Mabesa, R.; Dizon, E. Isolation and characterisation of selected lactic acid bacteria for improved processing of Nem chua, a traditional fermented meat from Vietnam. Benef. Microbes 2010, 1, 67–74. [Google Scholar] [CrossRef]
- Nacef, M.; Chevalier, M.; Chollet, S.; Drider, D.; Flahaut, C. MALDI-TOF mass spectrometry for the identification of lactic acid bacteria isolated from a French cheese: The Maroilles. Int. J. Food Microbiol. 2017, 247, 2–8. [Google Scholar] [CrossRef]
- Fukuda, K.; Shi, T.; Nagami, K.; Leo, F.; Nakamura, T.; Yasuda, K.; Senda, A.; Motoshima, H.; Urashima, T. Effects of carbohydrate source on physicochemical properties of the exopolysaccharide produced by Lactobacillus fermentum TDS030603 in a chemically defined medium. Carbohydr. Polym. 2010, 79, 1040–1045. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.; Zhang, C.; Luo, Y.; Zhang, H.; Yang, Z. Growth and exopolysaccharide production by Lactobacillus fermentum F6 in skim milk. Afr. J. Biotechnol. 2011, 10, 2080–2091. [Google Scholar] [CrossRef]
- Yang, Y.; Feng, F.; Zhou, Q.; Zhao, F.; Du, R.; Zhou, Z.; Han, Y. Isolation, purification and characterization of exopolysaccharide produced by Leuconostoc pseudomesenteroides YF32 from soybean paste. Int. J. Biol. Macromol. 2018, 114, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Górska, S.; Jachymek, W.; Rybka, J.; Strus, M.; Heczko, P.B.; Gamian, A. Structural and immunochemical studies of neutral exopolysaccharide produced by Lactobacillus johnsonii 142. Carbohydr. Res. 2010, 345, 108–114. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, C.; Li, D.; Zhao, Y.; Zhang, X.; Zeng, X.; Yang, Z.; Li, S. Antioxidant activity of an exopolysaccharide isolated from Lactobacillus plantarum C88. Int. J. Biol. Macromol. 2013, 54, 270–275. [Google Scholar] [CrossRef]
- Van Calsteren, M.-R.; Gagnon, F.; Nishimura, J.; Makino, S. Structure determination of the neutral exopolysaccharide produced by Lactobacillus delbrueckii subsp. bulgaricus OLL1073R-1. Carbohydr. Res. 2015, 413, 115–122. [Google Scholar] [CrossRef]
- Sánchez-Medina, I.; Gerwig, G.J.; Urshev, Z.L.; Kamerling, J.P. Structure of a neutral exopolysaccharide produced by Lactobacillus delbrueckii ssp. bulgaricus LBB. B26. Carbohydr. Res. 2007, 342, 2430–2439. [Google Scholar] [CrossRef]
- Gerwig, G.J.; Dobruchowska, J.M.; Shi, T.; Urashima, T.; Fukuda, K.; Kamerling, J.P. Structure determination of the exopolysaccharide of Lactobacillus fermentum TDS030603—A revision. Carbohydr. Res. 2013, 378, 84–90. [Google Scholar] [CrossRef] [Green Version]
- Saravanan, C.; Shetty, P.K.H. Isolation and characterization of exopolysaccharide from Leuconostoc lactis KC117496 isolated from idli batter. Int. J. Biol. Macromol. 2016, 90, 100–106. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.T.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Wang, K.; Li, W.; Rui, X.; Chen, X.; Jiang, M.; Dong, M. Structural characterization and bioactivity of released exopolysaccharides from Lactobacillus plantarum 70810. Int. J. Biol. Macromol. 2014, 67, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Cseke, L.J.; Herdy, J.R. Extraction/characterization of DNA. Methods Cell Biol. 2012, 112, 1–32. [Google Scholar] [CrossRef]
- Thi, T.T.V.; L#xEA; Trung Hiếu, L.T.L. Các thông số chất lượng của fucoidan và một số sản phẩm khác được phân lập từ rong mơ (sargassum) Thừa Thiên Huế. Hue Univ. J. Sci. (Hu Jos) 2013, 74, 141–150. [Google Scholar]
- Polak-Berecka, M.; Choma, A.; Waśko, A.; Górska, S.; Gamian, A.; Cybulska, J. Physicochemical characterization of exopolysaccharides produced by Lactobacillus rhamnosus on various carbon sources. Carbohydr. Polym. 2015, 117, 501–509. [Google Scholar] [CrossRef]
- Hussein, A.S.; Ibrahim, G.S.; Asker, M.M.S.; Mahmoud, M.G. Exopolysaccharide from Lactobacillus helveticus: Identification of chemical structure and effect on biscuit duality. Czech J. Food Sci. 2010, 28, 225–232. [Google Scholar] [CrossRef] [Green Version]
- Behare, P.V.; Singh, R.; Nagpal, R.; Rao, K. Exopolysaccharides producing Lactobacillus fermentum strain for enhancing rheological and sensory attributes of low-fat dahi. J. Food Sci. Technol. 2013, 50, 1228–1232. [Google Scholar] [CrossRef] [Green Version]
- Ale, E.C.; Perezlindo, M.J.; Pavón, Y.; Peralta, G.H.; Costa, S.; Sabbag, N.; Bergamini, C.; Reinheimer, J.A.; Binetti, A.G. Technological, rheological and sensory characterizations of a yogurt containing an exopolysaccharide extract from Lactobacillus fermentum Lf2, a new food additive. Food Res. Int. 2016, 90, 259–267. [Google Scholar] [CrossRef]
- Agrawal, P.K. NMR spectroscopy in the structural elucidation of oligosaccharides and glycosides. Phytochemistry 1992, 31, 3307–3330. [Google Scholar] [CrossRef]
- Harding, L.P.; Marshall, V.M.; Elvin, M.; Gu, Y.; Laws, A.P. Structural characterisation of a perdeuteriomethylated exopolysaccharide by NMR spectroscopy: Characterisation of the novel exopolysaccharide produced by Lactobacillus delbrueckii subsp. bulgaricus EU23. Carbohydr. Res. 2003, 338, 61–67. [Google Scholar] [CrossRef]
- Fontana, C.; Li, S.; Yang, Z.; Widmalm, G. Structural studies of the exopolysaccharide from Lactobacillus plantarum C88 using NMR spectroscopy and the program CASPER. Carbohydr. Res. 2015, 402, 87–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Górska-Frączek, S.; Sandström, C.; Kenne, L.; Paściak, M.; Brzozowska, E.; Strus, M.; Heczko, P.; Gamian, A. The structure and immunoreactivity of exopolysaccharide isolated from Lactobacillus johnsonii strain 151. Carbohydr. Res. 2013, 378, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Rani, R.P.; Anandharaj, M.; Ravindran, A.D. Characterization of a novel exopolysaccharide produced by Lactobacillus gasseri FR4 and demonstration of its in vitro biological properties. Int. J. Biol. Macromol. 2018, 109, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Zeng, Y.; Yang, M.; Chen, S.; He, L.; Ao, X.; Zou, L.; Liu, S. Production, purification and structural study of an exopolysaccharide from Lactobacillus plantarum BC-25. Carbohydr. Polym. 2016, 144, 205–214. [Google Scholar] [CrossRef]
- Sengun, I.Y.; Nielsen, D.S.; Karapinar, M.; Jakobsen, M. Identification of lactic acid bacteria isolated from Tarhana, a traditional Turkish fermented food. Int. J. Food Microbiol. 2009, 135, 105–111. [Google Scholar] [CrossRef]
- Goswami, G.; Bora, S.S.; Parveen, A.; Boro, R.C.; Barooah, M. Identification and functional properties of dominant lactic acid bacteria isolated from Kahudi, a traditional rapeseed fermented food product of Assam, India. J. Ethn. Foods 2017, 4, 187–197. [Google Scholar] [CrossRef]
- Nguyen, D.T.L.; Van Hoorde, K.; Cnockaert, M.; De Brandt, E.; De Bruyne, K.; Le, B.T.; Vandamme, P. A culture-dependent and -independent approach for the identification of lactic acid bacteria associated with the production of nem chua, a Vietnamese fermented meat product. Food Res. Int. 2013, 50, 232–240. [Google Scholar] [CrossRef]
- Torino, M.I.; Mozzi, F.; De Valdez, G.F. Exopolysaccharide biosynthesis by Lactobacillus helveticus ATCC 15807. Appl. Microbiol. Biotechnol. 2005, 68, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Yadav, V.; Prappulla, S.G.; Jha, A.; Poonia, A. A novel exopolysaccharide from probiotic Lactobacillus fermentum CFR 2195: Production, purification and characterization. Biotechnol. Bioinf. Bioeng. 2011, 1, 415–421. [Google Scholar]
- Freitas, F.; Alves, V.D.; Reis, M.A. Advances in bacterial exopolysaccharides: From production to biotechnological applications. Trends Biotechnol. 2011, 29, 388–398. [Google Scholar] [CrossRef]









| Sugar Residue | H-1 | H-2 | H-3 | H-4 | H-5 | H-6 | |
|---|---|---|---|---|---|---|---|
| A | →6)-β- D-glucopyranoside-(1→ | 4.87 | 3.53 | 3.62 | 3.49 | 3.28 | 3.51 |
| B | →3)-β-D- mannopyranoside-(1→ | 4.55 | 3.01 | 3.47 | 3.27 | 3.50 | 3.43 |
| Sugar Residue | C-1 | C-2 | C-3 | C-4 | C-5 | C-6 | |
| A | →6)-β- D-glucopyranoside-(1→ | 94.0 | 71.6 | 61.5 | 70.6 | 73.8 | 71.4 |
| B | →3)-β-D- mannopyranoside-(1→ | 94.1 | 77.2 | 73.2 | 67.1 | 67.4 | 61.5 |
| Sugar Residue | δH1 (ppm) | NOESY to | HMBC to | Connectivity | |
|---|---|---|---|---|---|
| (1→6)-β-D-glucopyranoside | A | 4.87 | A: H1 to B: H3 | ||
| 3.47 | 73.2 | A: H1 to B: C3 | |||
| (1→3)-β-D- mannopyranoside | B | 4.55 | B: H1 to A: H6 | ||
| 3.51 | 71.4 | B: H1 to A: C6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Do, T.B.T.; Tran, T.A.L.; Tran, T.V.T.; Le, T.H.; Jayasena, V.; Nguyen, T.H.C.; Nguyen, C.C.; Kim, S.Y.; Le, Q.V. Novel Exopolysaccharide Produced from Fermented Bamboo Shoot-Isolated Lactobacillus Fermentum. Polymers 2020, 12, 1531. https://doi.org/10.3390/polym12071531
Do TBT, Tran TAL, Tran TVT, Le TH, Jayasena V, Nguyen THC, Nguyen CC, Kim SY, Le QV. Novel Exopolysaccharide Produced from Fermented Bamboo Shoot-Isolated Lactobacillus Fermentum. Polymers. 2020; 12(7):1531. https://doi.org/10.3390/polym12071531
Chicago/Turabian StyleDo, Thi Bich Thuy, Thi Ai Luyen Tran, Thi Van Thi Tran, Trung Hieu Le, Vijay Jayasena, Thi Hong Chuong Nguyen, Chinh Chien Nguyen, Soo Young Kim, and Quyet Van Le. 2020. "Novel Exopolysaccharide Produced from Fermented Bamboo Shoot-Isolated Lactobacillus Fermentum" Polymers 12, no. 7: 1531. https://doi.org/10.3390/polym12071531
APA StyleDo, T. B. T., Tran, T. A. L., Tran, T. V. T., Le, T. H., Jayasena, V., Nguyen, T. H. C., Nguyen, C. C., Kim, S. Y., & Le, Q. V. (2020). Novel Exopolysaccharide Produced from Fermented Bamboo Shoot-Isolated Lactobacillus Fermentum. Polymers, 12(7), 1531. https://doi.org/10.3390/polym12071531