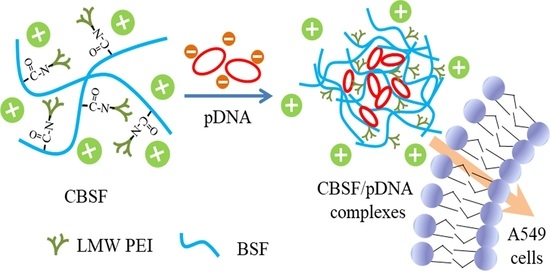
Polyethylenimine-Modified Bombyx mori Silk Fibroin as a Delivery Carrier of the ING4-IL-24 Coexpression Plasmid
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of Silk Fibroin Solution
2.2. Preparation of Cationized Silk Fibroin Solution
2.3. Characterization of CBSF
2.4. Construction of Recombinant Plasmids Coexpressing ING4 and IL-24 Double Genes
2.5. Preparation of CBSF/pDNA and PEI/pDNA Complexes
2.6. Gel Retardation Assay
2.7. Characterization of CBSF/pDNA Complexes
2.8. In Vitro Gene Transfection
2.9. Evaluation of Cell Viability
2.10. Transfection Efficiency Evaluation
2.11. Statistical Analysis
3. Results
3.1. Characterization of CBSF
3.2. Characterization of CBSF
3.3. In Vitro Transfection of A549 and WI-38 Cells with CBSF/pDNA Complexes
3.4. Transfection Efficiency of Complexes Transfect A549 Cells
3.5. Effects of CBSF/pDNA Complexes on the Proliferation of A549 and WI-38 Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laksee, S.; Supachettapun, C.; Muangsin, N.; Lertsarawut, P.; Rattanawongwiboon, T.; Sricharoen, P.; Limchoowong, N.; Chutimasakul, T.; Kwamman, T.; Hemvichian, K. Targeted gold nanohybrids functionalized with folate-hydrophobic-quaternized pullulan delivering camptothecin for enhancing hydrophobic anticancer drug efficacy. Polymers 2021, 13, 2670. [Google Scholar] [CrossRef]
- Engel, J.; Lategahn, J.; Rauh, D. Hope and disappointment: Covalent inhibitors to overcome drug resistance in non-small cell lung cancer. ACS Med. Chem. Lett. 2016, 7, 2–5. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Mohammed, K.A.; Nasreen, N. Nanoparticle-based targeted gene therapy for lung cancer. Nanoparticles Lung Cancer 2016, 6, 1118–1134. [Google Scholar]
- Xu, M.; Xie, Y.; Sheng, W.; Miao, J.; Yang, J. Adenovirus-mediated ING4 gene transfer in osteosarcoma suppresses tumor growth via induction of apoptosis and inhibition of tumor angiogenesis. Technol. Cancer Res. Treat. 2014, 14, 369–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nozell, S.; Laver, T.; Moseley, D.; Nowoslawski, L.; De Vos, M.; Atkinson, G.P.; Harrison, K.; Nabors, L.B.; Benveniste, E.N. The ING4 tumor suppressor attenuates NF-κB activity at the promoters of target genes. Mol. Cell Biol. 2008, 28, 6632–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Zhu, Y.; Zhang, H.; Li, L.; He, P.; Xia, H.; Zhang, Y.; Mao, C. Delivery of inhibitor of growth 4 (ING4) gene significantly inhibits proliferation and invasion and promotes apoptosis of human osteosarcoma cells. Sci. Rep. 2014, 4, 7380–7389. [Google Scholar] [CrossRef] [Green Version]
- Panneerselvam, J.; Srivastava, A.; Mehta, M.; Chen, A.; Zhao, Y.D.; Munshi, A.; Ramesh, R. IL-24 inhibits lung cancer growth by suppressing GLI1 and inducing DNA damage. Cancers 2019, 11, 1879. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Emdad, L.; Sauane, M.; Lebedeva, I.V.; Sarkar, D.; Gupta, P.; James, C.D.; Randolph, A.; Valerie, K.; Walter, M.R.; et al. Unique aspects of mda-7/IL-24 antitumor bystander activity: Establishing a role for secretion of MDA-7/IL-24 protein by normal cells. Oncogene 2005, 24, 7552–7566. [Google Scholar] [CrossRef] [Green Version]
- Qu, J.; Wang, W.; Feng, Y.; Niu, L.; Li, M.; Yang, J.; Xie, Y. Cationic antheraea pernyi silk fibroin-modified adenovirus-mediated ING4 and IL-24 dual gene coexpression vector suppresses the growth of hepatoma carcinoma cells. Int. J. Nanomed. 2019, 14, 9745–9761. [Google Scholar] [CrossRef] [Green Version]
- Kuzmich, A.; Rakitina, O.; Didych, D.; Potapov, V.; Zinovyeva, M.; Alekseenko, I.; Sverdlov, E. Novel histone-based DNA carrier targeting cancer-associated fibroblasts. Polymers 2020, 12, 1695. [Google Scholar] [CrossRef]
- Santana-Armas, M.L.; Tros de Ilarduya, C. Strategies for cancer gene-delivery improvement by non-viral vectors. Int. J. Pharm. 2021, 596, 120291–120299. [Google Scholar] [CrossRef] [PubMed]
- Wahane, A.; Waghmode, A.; Kapphahn, A.; Dhuri, K.; Gupta, A.; Bahal, R. Role of lipid-based and polymer-based non-viral vectors in nucleic acid delivery for next-generation gene therapy. Molecules 2020, 25, 2866. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.Y.; Zhan, Y.R.; Zhang, J.; Luan, C.R.; Wang, B.; Yu, X.Q. Aromatic modification of low molecular weight PEI for enhanced gene delivery. Polymers 2017, 9, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babaei, M.; Eshghi, H.; Abnous, K.; Rahimizadeh, M.; Ramezani, M. Promising gene delivery system based on polyethylenimine-modified silica nanoparticles. Cancer Gene Ther. 2017, 24, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, S.; Chen, T.; Xian, W.; Zhang, H.; Wu, L.; Zhu, W.; Zeng, Q. Nanoscale cationic micelles of amphiphilic copolymers based on star-shaped PLGA and PEI cross-linked PEG for protein delivery application. J. Mater. Sci. Mater. Med. 2019, 30, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Hao, X.; Wang, H.; Guo, J.; Ren, X.K.; Xia, S.; Zhang, W.; Feng, Y. Multifunctional REDV-G-TAT-G-NLS-Cys peptide sequence conjugated gene carriers to enhance gene transfection efficiency in endothelial cells. Colloids Surf. B Biointerfaces 2019, 184, 110510–110520. [Google Scholar] [CrossRef]
- Kuo, W.T.; Huang, H.Y.; Chou, M.J.; Wu, M.C.; Huang, Y.Y. Surface modification of gelatin nanoparticles with polyethylenimine as gene vector. J. Nanomater. 2011, 2011, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Song, J.; Liang, X.; Yin, Y.; Zuo, T.; Chen, D.; Shen, Q. Hyaluronic acid-modified cationic nanoparticles overcome enzyme CYP1B1-mediated breast cancer multidrug resistance. Nanomedicine 2019, 14, 447–464. [Google Scholar] [CrossRef]
- Clima, L.; Craciun, B.F.; Gavril, G.; Pinteala, M. Tunable composition of dynamic non-viral vectors over the DNA polyplex formation and nucleic acid transfection. Polymers 2019, 11, 1313. [Google Scholar] [CrossRef] [Green Version]
- Sharmeen, S.; Rahman, A.M.; Lubna, M.M.; Salem, K.S.; Islam, R.; Khan, M.A. Polyethylene glycol functionalized carbon nanotubes/gelatin-chitosan nanocomposite: An approach for significant drug release. Bioact. Mater. 2018, 3, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Xu, H.; Lan, Y.; Zhu, Q.; Liu, Y.; Huang, S.; Shi, S.; Hancharou, A.; Tang, B.; Guo, R. Preparation and characterisation of a novel silk fibroin/hyaluronic acid/sodium alginate scaffold for skin repair. Int. J. Biol. Macromol. 2019, 130, 58–67. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Q.; Li, H.; Wei, Q.; Zhao, X.; Chen, F. Elastin-like polypeptide modified silk fibroin porous scaffold promotes osteochondral repair. Bioact. Mater. 2021, 6, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Norouzi, P.; Motasadizadeh, H.; Atyabi, F.; Dinarvand, R.; Gholami, M.; Farokhi, M.; Shokrgozar, M.A.; Mottaghitalab, F. Combination therapy of breast cancer by codelivery of doxorubicin and survivin siRNA using polyethylenimine modified silk fibroin nanoparticles. ACS Biomater. Sci. Eng. 2021, 7, 1074–1087. [Google Scholar] [CrossRef] [PubMed]
- Tomeh, M.A.; Hadianamrei, R.; Zhao, X. Silk fibroin as a functional biomaterial for drug and gene delivery. Pharmaceutics 2019, 11, 494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zheng, Z.; Gong, H.; Liu, M.; Guo, S.; Li, G.; Wang, X.; Kaplan, D.L. DNA preservation in silk. Biomater. Sci. 2017, 5, 1279–1292. [Google Scholar] [CrossRef]
- Farokhi, M.; Mottaghitalab, F.; Reis, R.L.; Ramakrishna, S.; Kundu, S.C. Functionalized silk fibroin nanofibers as drug carriers: Advantages and challenges. J. Control Release 2020, 321, 324–347. [Google Scholar] [CrossRef]
- Luo, Z.; Li, J.; Qu, J.; Sheng, W.; Yang, J.; Li, M. Cationized bombyx mori silk fibroin as a delivery carrier of the VEGF165-Ang-1 coexpression plasmid for dermal tissue regeneration. J. Mater. Chem. B 2019, 7, 80–94. [Google Scholar] [CrossRef]
- Niu, L.; Shi, M.; Feng, Y.; Sun, X.; Wang, Y.; Cheng, Z.; Li, M. The interactions of quantum dot-labeled silk fibroin micro/nanoparticles with cells. Materials 2020, 13, 3372. [Google Scholar] [CrossRef]
- Kajiwara, S.; Komatsu, K.; Yamada, R.; Matsumoto, T.; Yasuda, M.; Ogino, H. Improvement of the organic solvent stability of a commercial lipase by chemical modification with dextran. Biochem. Eng. J. 2019, 142, 1–6. [Google Scholar] [CrossRef]
- Xie, Y.; Lv, H.; Sheng, W.; Miao, J.; Xiang, J.; Yang, J. Synergistic tumor suppression by adenovirus-mediated inhibitor of growth 4 and interleukin-24 gene cotransfer in hepatocarcinoma cells. Cancer Biother. Radiopharm. 2011, 26, 681–695. [Google Scholar] [CrossRef]
- Liu, H.; Li, K.; Xu, L.; Wu, D. Bilayered near-infrared fluorescent nanoparticles based on low molecular weight PEI for tumor-targeted in vivo imaging. J. Nanopart. Res. 2014, 16, 2784–2795. [Google Scholar] [CrossRef]
- Dong, H.; Ding, L.; Yan, F.; Ji, H.; Ju, H. The use of polyethylenimine-grafted graphene nanoribbon for cellular delivery of locked nucleic acid modified molecular beacon for recognition of microRNA. Biomaterials 2011, 32, 3875–3882. [Google Scholar] [CrossRef]
- Byram, P.K.; Sunka, K.C.; Barik, A.; Kaushal, M.; Dhara, S.; Chakravorty, N. Biomimetic silk fibroin and xanthan gum blended hydrogels for connective tissue regeneration. Int. J. Biol. Macromol. 2020, 165, 874–882. [Google Scholar] [CrossRef]
- Shao, J.; Zheng, J.; Liu, J.; Carr, C.M. Fourier transform Raman and Fourier transform infrared spectroscopy studies of silk fibroin. J. Appl. Polym. Sci. 2005, 96, 1999–2004. [Google Scholar] [CrossRef]
- Nurhasni, H.; Cao, J.; Choi, M.; Kim, I.; Lee, B.L.; Jung, Y.; Yoo, J.W. Nitric oxide-releasing poly (lactic-co-glycolic acid)-polyethylenimine nanoparticles for prolonged nitric oxide release, antibacterial efficacy, and in vivo wound healing activity. Int. J. Nanomed. 2015, 10, 3065–3080. [Google Scholar]
- Zainuddin; Le, T.T.; Park, Y.; Chirila, T.V.; Halley, P.J.; Whittaker, A.K. The behavior of aged regenerated Bombyx mori silk fibroin solutions studied by 1H NMR and rheology. Biomaterials 2008, 29, 4268–4274. [Google Scholar] [CrossRef] [PubMed]
- Ping, Y.; Hu, Q.; Tang, G.; Li, J. FGFR-targeted gene delivery mediated by supramolecular assembly between β-cyclodextrin-crosslinked PEI and redox-sensitive PEG. Biomaterials 2013, 34, 6482–6494. [Google Scholar] [CrossRef]
- Guo, S.; Liang, Y.; Liu, L.; Yin, M.; Wang, A.; Sun, K.; Li, Y.; Shi, Y. Research on the fate of polymeric nanoparticles in the process of the intestinal absorption based on model nanoparticles with various characteristics: Size, surface charge and pro-hydrophobics. J. Nanobiotechnol. 2021, 19, 32–52. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Zhao, J.; Su, B.; Rukh, S.; Guo, J.; Ren, X.K.; Xia, S.; Zhang, W.; Feng, Y. Redox stimulus disulfide conjugated polyethyleneimine as a shuttle for gene transfer. J. Mater. Sci. Mater. Med. 2020, 31, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.R.; Zhang, J.; Zhang, J.H.; Xiao, Y.P.; He, X.; Liu, Y.H.; Yu, X.Q. Amino acid-linked low molecular weight polyethylenimine for Improved gene delivery and biocompatibility. Molecules 2020, 25, 975. [Google Scholar] [CrossRef] [Green Version]
- Syga, M.I.; Nicoli, E.; Kohler, E.; Shastri, V.P. Albumin incorporation in polyethylenimine-DNA polyplexes influences transfection efficiency. Biomacromolecules 2016, 17, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Forest, V.; Pourchez, J. Preferential binding of positive nanoparticles on cell membranes is due to electrostatic interactions: A too simplistic explanation that does not take into account the nanoparticle protein corona. Mater. Sci. Eng. C 2017, 70, 889–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Guo, H.; Liu, L.; Liu, Y.; Xie, L. Size-dependent cellular uptake and localization profiles of silver nanoparticles. Int. J. Nanomed. 2019, 14, 4247–4259. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Wang, Q.; Zhang, Z.; Gong, T.; Sun, X. Cationic bovine serum albumin based self-assembled nanoparticles as siRNA delivery vector for treating lung metastatic cancer. Small 2014, 10, 524–535. [Google Scholar] [CrossRef]
- Wu, D.; Zhang, Y.; Xu, X.; Guo, T.; Xie, D.; Zhu, R.; Chen, S.; Ramakrishna, S.; He, L. RGD/TAT-functionalized chitosan-graft-PEI-PEG gene nanovector for sustained delivery of NT-3 for potential application in neural regeneration. Acta Biomater. 2018, 72, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Xing, H.; Sun, Y.; Feng, S.; Wang, S. Non-small cell lung cancer combination therapy: Hyaluronic acid modified, epidermal growth factor receptor targeted, pH sensitive lipid-polymer hybrid nanoparticles for the delivery of erlotinib plus bevacizumab. Biomed. Pharmacother. 2020, 125, 109861. [Google Scholar] [CrossRef]
- Tan, S.; Wang, G. Lung cancer targeted therapy: Folate and transferrin dual targeted, glutathione responsive nanocarriers for the delivery of cisplatin. Biomed Pharmacother. 2018, 102, 55–63. [Google Scholar] [CrossRef]
- Kunath, K. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: Comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J. Control Release 2003, 89, 113–125. [Google Scholar] [CrossRef]
- Tian, H.; Lin, L.; Jiao, Z.; Guo, Z.; Chen, J.; Gao, S.; Zhu, X.; Chen, X. Polylysine-modified polyethylenimine inducing tumor apoptosis as an efficient gene carrier. J. Control Release 2013, 172, 410–418. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, L.; Chen, G.; Feng, Y.; Liu, X.; Pan, P.; Huang, L.; Guo, Y.; Li, M. Polyethylenimine-Modified Bombyx mori Silk Fibroin as a Delivery Carrier of the ING4-IL-24 Coexpression Plasmid. Polymers 2021, 13, 3592. https://doi.org/10.3390/polym13203592
Niu L, Chen G, Feng Y, Liu X, Pan P, Huang L, Guo Y, Li M. Polyethylenimine-Modified Bombyx mori Silk Fibroin as a Delivery Carrier of the ING4-IL-24 Coexpression Plasmid. Polymers. 2021; 13(20):3592. https://doi.org/10.3390/polym13203592
Chicago/Turabian StyleNiu, Longxing, Guo Chen, Yanfei Feng, Xueping Liu, Peng Pan, Linling Huang, Ying Guo, and Mingzhong Li. 2021. "Polyethylenimine-Modified Bombyx mori Silk Fibroin as a Delivery Carrier of the ING4-IL-24 Coexpression Plasmid" Polymers 13, no. 20: 3592. https://doi.org/10.3390/polym13203592
APA StyleNiu, L., Chen, G., Feng, Y., Liu, X., Pan, P., Huang, L., Guo, Y., & Li, M. (2021). Polyethylenimine-Modified Bombyx mori Silk Fibroin as a Delivery Carrier of the ING4-IL-24 Coexpression Plasmid. Polymers, 13(20), 3592. https://doi.org/10.3390/polym13203592