Fluorescent BAPAD Dendrimeric Antigens Are Efficiently Internalized by Human Dendritic Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Structural Characterization
2.3. General Procedure for Amide Formation
2.4. General Procedure for Azide Reduction
2.5. Synthesis of Compound 9
2.6. Synthesis of Compound 11
2.7. Synthesis of Compound 1
2.8. Synthesis of Compound 13
2.9. Synthesis of Compound 2
2.10. Fluorescence Characterization
2.11. Molecular Dynamics Simulations
2.12. Generation of Monocyte-Derived DCs
2.13. Flow Cytometry-Based Detection of Compounds 1 and 2 in DCs
2.14. Cell Toxicity Analyses
3. Results and Discussion
3.1. Synthesis
3.2. Fluorescent Properties Study
3.3. Theoretical Calculations of Compounds 1 and 2
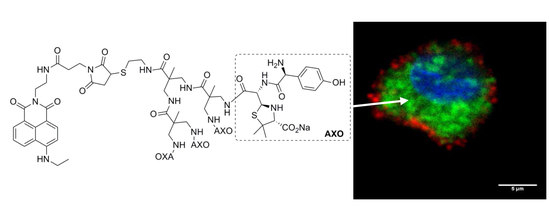
3.4. Biomedical Applications of Compounds 1 and 2
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ornelas, C. Brief timelapse on dendrimer chemistry: Advances, limitations, and expectations. Macromol. Chem. Phys. 2016, 217, 149–174. [Google Scholar] [CrossRef]
- Caminade, A.-M.; Ouali, A.; Laurent, R.; Turrin, C.-O.; Majoral, J.-P. Coordination chemistry with phosphorus dendrimers. Applications as catalysts, for materials, and in biology. Coord. Chem. Rev. 2016, 308, 478–497. [Google Scholar] [CrossRef]
- Bahadır, E.B.; Sezgintürk, M.K. Poly(amidoamine) (PAMAM): An emerging material for electrochemical bio(sensing) applications. Talanta 2016, 148, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sanchez, A.J.; Mesa-Antunez, P.; Barbero, N.; Collado, D.; Vida, Y.; Najera, F.; Perez-Inestrosa, E. Synthesis of all-aliphatic polyamide dendrimers based on a 3,3′-diaminopivalic acid scaffold. Polym. Chem. 2015, 6, 3031–3038. [Google Scholar] [CrossRef]
- Backlund, C.J.; Worley, B.V.; Schoenfisch, M.H. Anti-biofilm action of nitric oxide-releasing alkyl-modified poly(amidoamine) dendrimers against Streptococcus mutans. Acta Biomater. 2016, 29, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Kesharwani, P.; Banerjee, S.; Gupta, U.; Mohd Amin, M.C.I.; Padhye, S.; Sarkar, F.H.; Iyer, A.K. PAMAM dendrimers as promising nanocarriers for RNAi therapeutics. Mater. Today 2015, 18, 565–572. [Google Scholar] [CrossRef]
- Hashemi, M.; Tabatabai, S.M.; Parhiz, H.; Milanizadeh, S.; Amel Farzad, S.; Abnous, K.; Ramezani, M. Gene delivery efficiency and cytotoxicity of heterocyclic amine-modified PAMAM and PPI dendrimers. Mater. Sci. Eng. C 2016, 61, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Hua, W.; Zhang, Y.; Li, C.; Xue, W.; Yin, J.; Liu, Z.; Qiu, X. Effect of poly(amidoamine) dendrimers on the structure and activity of immune molecules. Biochim. Biophys. Acta BBA Gen. Subj. 2015, 1850, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Mammen, M.; Choi, S.-K.; Whitesides, G.M. Polyvalent interactions in biological systems: Implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998, 37, 2754–2794. [Google Scholar] [CrossRef]
- Naisbitt, D.J.; Gordon, S.; Pirmohamed, M.; Park, B. Immunological principles of adverse drug reactions: The initiation and propagation of immune responses elicited by drug treatment. Drug Saf. 2000, 23, 483–507. [Google Scholar] [CrossRef] [PubMed]
- Antunez, C.; Blanca-Lopez, N.; Torres, M.J.; Mayorga, C.; Perez-Inestrosa, E.; Montañez, M.I.; Fernandez, T.; Blanca, M. Immediate allergic reactions to cephalosporins: Evaluation of cross-reactivity with a panel of penicillins and cephalosporins. J. Allergy Clin. Immunol. 2006, 117, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Montañez, M.I.; Mayorga, C.; Torres, M.J.; Ariza, A.; Blanca, M.; Perez-Inestrosa, E. Synthetic Approach to gain insight into antigenic determinants of cephalosporins: In vitro studies of chemical structure−IgE molecular recognition relationships. Chem. Res. Toxicol. 2011, 24, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Vida, Y.; Montañez, M.I.; Collado, D.; Najera, F.; Ariza, A.; Blanca, M.; Torres, M.J.; Mayorga, C.; Perez-Inestrosa, E. Dendrimeric antigen–silica particle composites: An innovative approach for IgE quantification. J. Mater. Chem. B 2013, 1, 3044–3050. [Google Scholar] [CrossRef]
- Montañez, M.I.; Najera, F.; Perez-Inestrosa, E. NMR Studies and molecular dynamic simulation of synthetic dendritic antigens. Polymers 2011, 3, 1533–1553. [Google Scholar] [CrossRef]
- Li, Y.; Guo, K.; Su, H.; Li, X.; Feng, X.; Wang, Z.; Zhang, W.; Zhu, S.; Wesdemiotis, C.; Cheng, S.Z.D.; et al. Tuning “thiol-ene” reactions toward controlled symmetry breaking in polyhedral oligomeric silsesquioxanes. Chem. Sci. 2014, 5, 1046–1053. [Google Scholar] [CrossRef]
- Hull, E.A.; Livanos, M.; Miranda, E.; Smith, M.E.B.; Chester, K.A.; Baker, J.R. Homogeneous bispecifics by disulfide bridging. Bioconjug. Chem. 2014, 25, 1395–1401. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Wang, Y.; Ma, Z.; Wang, Z.; Wei, Y.; Jia, X. A poly(amidoamine) dendrimer-based nanocarrier conjugated with Angiopep-2 for dual-targeting function in treating glioma cells. Polym. Chem. 2016, 7, 715–721. [Google Scholar] [CrossRef]
- Wang, K.; Hu, Q.; Zhu, W.; Zhao, M.; Ping, Y.; Tang, G. Structure-Invertible nanoparticles for triggered co-delivery of nucleic acids and hydrophobic drugs for combination cancer therapy. Adv. Funct. Mater. 2015, 25, 3380–3392. [Google Scholar] [CrossRef]
- Soler, M.; Mesa-Antunez, P.; Estevez, M.-C.; Ruiz-Sanchez, A.J.; Otte, M.A.; Sepulveda, B.; Collado, D.; Mayorga, C.; Torres, M.J.; Perez-Inestrosa, E.; et al. Highly sensitive dendrimer-based nanoplasmonic biosensor for drug allergy diagnosis. Biosens. Bioelectron. 2015, 66, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Mckenna, M.; Grabchev, I.; Bosch, P. The synthesis of a novel 1,8-naphthalimide based PAMAM-type dendron and its potential for light-harvesting. Dyes Pigments 2009, 81, 180–186. [Google Scholar] [CrossRef]
- Song, H.Y.; Ngai, M.H.; Song, Z.Y.; MacAry, P.A.; Hobley, J.; Lear, M.J. Practical synthesis of maleimides and coumarin-linked probes for protein and antibody labelling via reduction of native disulfides. Org. Biomol. Chem. 2009, 7, 3400–3406. [Google Scholar] [CrossRef] [PubMed]
- Maingi, V.; Jain, V.; Bharatam, P.V.; Maiti, P.K. Dendrimer building toolkit: Model building and characterization of various dendrimer architectures. J. Comput. Chem. 2012, 33, 1997–2011. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Darden, T.A.; Cheatham, T.E., III; Simmerling, C.L.; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.M.; et al. AMBER 12; AMBER; University of California: San Francisco, CA, USA, 2012. [Google Scholar]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Maj, A.M.; Suisse, I.; Pinault, N.; Robert, N.; Agbossou-Niedercorn, F. Efficient catalytic hydrogenation of N-unsubstituted cyclic imides to cyclic amines. ChemCatChem 2014, 6, 2621–2625. [Google Scholar] [CrossRef]
- Tomalia, D.; Huang, B.; Swanson, D.; Brothers, H.; Klimash, J. Structure control within poly(amidoamine) dendrimers: Size, shape and regio-chemical mimicry of globular proteins. Tetrahedron 2003, 59, 3799–3813. [Google Scholar] [CrossRef]
- Schotten, C. Ueber die Oxydation des Piperidins. Berichte Dtsch. Chem. Ges. 1884, 17, 2544–2547. [Google Scholar] [CrossRef]
- Baumann, E. Ueber eine einfache Methode der Darstellung von Benzoësäureäthern. Berichte Dtsch. Chem. Ges. 1886, 19, 3218–3222. [Google Scholar] [CrossRef]
- Staudinger, H.; Meyer, J. Über neue organische Phosphorverbindungen III. Phosphinmethylenderivate und Phosphinimine. Helv. Chim. Acta 1919, 2, 635–646. [Google Scholar] [CrossRef]
- Pilard, S.; Vaultier, M. A stereospecific synthesis of (±) α-conhydrine and (±) β-conhydrine. Tetrahedron Lett. 1984, 25, 1555–1556. [Google Scholar] [CrossRef]
- Lagunas, A.; Castaño, A.G.; Artés, J.M.; Vida, Y.; Collado, D.; Pérez-Inestrosa, E.; Gorostiza, P.; Claros, S.; Andrades, J.A.; Samitier, J. Large-scale dendrimer-based uneven nanopatterns for the study of local arginine-glycine-aspartic acid (RGD) density effects on cell adhesion. Nano Res. 2014, 7, 399–409. [Google Scholar] [CrossRef]
- Hung, W.-C.; Cherng, J.-Y. Maleimide-functionalized PEI600 grafted polyurethane: synthesis, nano-complex formation with DNA and thiol-conjugation of the complexes for dual DNA transfection. Polymers 2015, 7, 2131–2145. [Google Scholar] [CrossRef]
- Brouwer, A.M. Standards for photoluminescence quantum yield measurements in solution (IUPAC Technical Report). Pure Appl. Chem. 2011, 83, 2213–2228. [Google Scholar] [CrossRef]
- Guy, J.; Caron, K.; Dufresne, S.; Michnick, S.W.; Skene, W.G.; Keillor, J.W. Convergent preparation and photophysical characterization of dimaleimide dansyl fluorogens: Elucidation of the maleimide fluorescence quenching mechanism. J. Am. Chem. Soc. 2007, 129, 11969–11977. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Samanta, A. Influence of the structure of the amino group and polarity of the medium on the photophysical behavior of 4-amino-1,8-naphthalimide derivatives. J. Phys. Chem. A 2002, 106, 4763–4771. [Google Scholar] [CrossRef]
- Wehner, R.; Dietze, K.; Bachmann, M.; Schmitz, M. The bidirectional crosstalk between human dendritic cells and natural killer cells. J. Innate Immun. 2011, 3, 258–263. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Pena, R.; Lopez, S.; Mayorga, C.; Antunez, C.; Fernandez, T.; Torres, M.J.; Blanca, M. Potential involvement of dendritic cells in delayed-type hypersensitivity reactions to β-lactams. J. Allergy Clin. Immunol. 2006, 118, 949–956. [Google Scholar] [CrossRef] [PubMed]















| Compound | ΦF | τ | ε |
|---|---|---|---|
| 9 | 0.62 1 | 10 1 | – |
| 11 | 0.25 1 | 4 1 | – |
| 1 | 0.62 1 0.18 2 | 10 1 4 2 | 7738 2 |
| 2 | 0.57 1 0.12 2 | 10 1 4 2 | 13,602 2 |
| Dendron | Ix/Iy | Ix/Iz | δ | Rg (Å) | RSASA (Å) |
|---|---|---|---|---|---|
| 1 | 1.18 ± 0.04 | 2.68 ± 0.35 | 0.064 ± 0.014 | 5.94 ± 0.18 | 7.47 |
| 2 | 1.30 ± 0.07 | 1.64 ± 0.17 | 0.021 ± 0.007 | 8.56 ± 0.41 | 11.49 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mesa-Antunez, P.; Collado, D.; Vida, Y.; Najera, F.; Fernandez, T.; Torres, M.J.; Perez-Inestrosa, E. Fluorescent BAPAD Dendrimeric Antigens Are Efficiently Internalized by Human Dendritic Cells. Polymers 2016, 8, 111. https://doi.org/10.3390/polym8040111
Mesa-Antunez P, Collado D, Vida Y, Najera F, Fernandez T, Torres MJ, Perez-Inestrosa E. Fluorescent BAPAD Dendrimeric Antigens Are Efficiently Internalized by Human Dendritic Cells. Polymers. 2016; 8(4):111. https://doi.org/10.3390/polym8040111
Chicago/Turabian StyleMesa-Antunez, Pablo, Daniel Collado, Yolanda Vida, Francisco Najera, Tahia Fernandez, Maria Jose Torres, and Ezequiel Perez-Inestrosa. 2016. "Fluorescent BAPAD Dendrimeric Antigens Are Efficiently Internalized by Human Dendritic Cells" Polymers 8, no. 4: 111. https://doi.org/10.3390/polym8040111
APA StyleMesa-Antunez, P., Collado, D., Vida, Y., Najera, F., Fernandez, T., Torres, M. J., & Perez-Inestrosa, E. (2016). Fluorescent BAPAD Dendrimeric Antigens Are Efficiently Internalized by Human Dendritic Cells. Polymers, 8(4), 111. https://doi.org/10.3390/polym8040111