Multifunctional Polymer Nanoparticles for Dual Drug Release and Cancer Cell Targeting
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of Methacrylic Acid N-hydroxysuccinimide Ester (MAA–NHS)
2.3. Synthesis of Biotin N-hydroxysuccinimide Ester (Biotin–NHS)
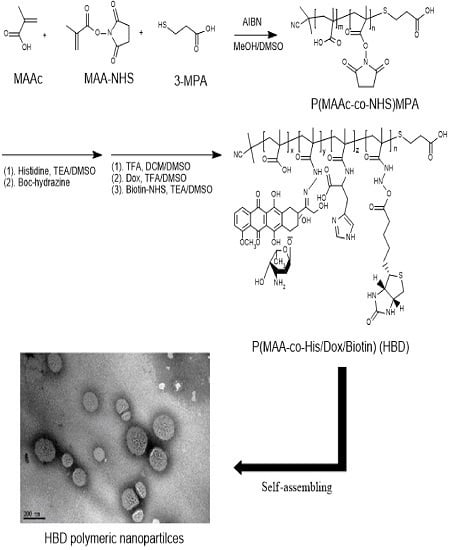
2.4. Synthesis of BD and HBD Copolymers
2.5. Preparation of IMQ-Loaded Nanoparticles
2.6. Measurement of Drug Release in Various pH Conditions
2.7. Cellular Uptake Analysis by Flow Cytometry
3. Results and Discussion
3.1. Characterizations of BD and HBD Copolymers
3.2. Characterizations of BD and HBD Nanoparticles
3.3. In Vitro Drug Release Behaviors of BD and HBD Nanoparticles
3.4. In Vitro Cellular Uptake
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tosi, G.; Costantino, L.; Ruozi, B.; Forni, F.; Vandelli, M.A. Polymeric nanoparticles for the drug delivery to the central nervous system. Expert Opin. Drug Deliv. 2008, 5, 155–174. [Google Scholar] [CrossRef] [PubMed]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.F.; Wu, P.; Jiang, Z.H.; Wei, X.Y. Synthesis and tumor cell growth inhibitory activity of biotinylated annonaceous acetogenins. Eur. J. Med. Chem. 2014, 71, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Kularatne, S.A.; Low, P.S. Targeting of nanoparticles: Folate receptor. Methods Mol. Biol. 2010, 624, 249–265. [Google Scholar] [PubMed]
- Daniels, T.R.; Bernabeu, E.; Rodriguez, J.A.; Patel, S.; Kozman, M.; Chiappetta, D.A.; Holler, E.; Ljubimova, J.Y.; Helguera, G.; Penichet, M.L. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim. Biophys. Acta 2012, 1820, 291–317. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Niestroj, M.; Yuan, D.; Chang, S.; Chen, J. Treating cancer stem cells and cancer metastasis using glucose-coated gold nanoparticles. Int. J. Nanomed. 2015, 10, 2065–2077. [Google Scholar]
- MacEwan, S.R.; Callahan, D.J.; Chilkoti, A. Stimulus-responsive macromolecules and nanoparticles for cancer drug delivery. Nanomedicine 2010, 5, 793–806. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Huang, L. Exploring the tumor microenvironment with nanoparticles. Cancer Treat. Res. 2015, 166, 193–226. [Google Scholar] [PubMed]
- Rosen, T.; Harting, M.; Gibson, M. Treatment of bowen’s disease with topical 5% imiquimod cream: Retrospective study. Dermatol. Surg. 2007, 33, 427–431, discussion 431–422. [Google Scholar] [CrossRef] [PubMed]
- Vasilakos, J.P.; Tomai, M.A. The use of toll-like receptor 7/8 agonists as vaccine adjuvants. Expert Rev. Vaccines 2013, 12, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Gambara, G.; De Cesaris, P.; De Nunzio, C.; Ziparo, E.; Tubaro, A.; Filippini, A.; Riccioli, A. Toll-like receptors in prostate infection and cancer between bench and bedside. J. Cell. Mol. Med. 2013, 17, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Seth, A.; Heo, M.B.; Lim, Y.T. Poly (γ-glutamic acid) based combination of water-insoluble paclitaxel and tlr7 agonist for chemo-immunotherapy. Biomaterials 2014, 35, 7992–8001. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Sorkin, A.; Von Zastrow, M. Signal transduction and endocytosis: Close encounters of many kinds. Nat. Rev. Mol. Cell Biol. 2002, 3, 600–614. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Qiu, L.; Chen, Q.; Hao, T.; Qiao, M.; Zhao, H.; Zhang, J.; Hu, H.; Zhao, X.; Chen, D.; et al. pH-sensitive nanoparticles of poly(l-histidine)-poly(lactide-co-glycolide)-tocopheryl polyethylene glycol succinate for anti-tumor drug delivery. Acta Biomater. 2015, 11, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.M.; Bae, Y.H.; Jo, W.H. pH-induced micelle formation of poly(histidine-co-phenylalanine)-block-poly(ethylene glycol) in aqueous media. Macromol. Biosci. 2005, 5, 1118–1124. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhao, X.; Chen, J.; Chen, J.; Kuznetsova, L.; Wong, S.S.; Ojima, I. Mechanism-based tumor-targeting drug delivery system. Validation of efficient vitamin receptor-mediated endocytosis and drug release. Bioconj. Chem. 2010, 21, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Gurcel, C.; Vercoutter-Edouart, A.S.; Fonbonne, C.; Mortuaire, M.; Salvador, A.; Michalski, J.C.; Lemoine, J. Identification of new O-glcnac modified proteins using a click-chemistry-based tagging. Anal. Bioanal. Chem. 2008, 390, 2089–2097. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Watanabe, R.; Choyke, P.L. Improving conventional enhanced permeability and retention (EPR) effects; what is the appropriate target? Theranostics 2013, 4, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Sandberg, A.; Heckert, E.; Self, W.; Seal, S. Protein adsorption and cellular uptake of cerium oxide nanoparticles as a function of zeta potential. Biomaterials 2007, 28, 4600–4607. [Google Scholar] [CrossRef] [PubMed]
- Schon, M.; Bong, A.B.; Drewniok, C.; Herz, J.; Geilen, C.C.; Reifenberger, J.; Benninghoff, B.; Slade, H.B.; Gollnick, H.; Schon, M.P. Tumor-selective induction of apoptosis and the small-molecule immune response modifier imiquimod. J. Natl. Cancer Inst. 2003, 95, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Prins, R.M.; Craft, N.; Bruhn, K.W.; Khan-Farooqi, H.; Koya, R.C.; Stripecke, R.; Miller, J.F.; Liau, L.M. The tlr-7 agonist, imiquimod, enhances dendritic cell survival and promotes tumor antigen-specific T cell priming: Relation to central nervous system antitumor immunity. J. Immunol. 2006, 176, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Heffernan, M.J.; Murthy, N. Polyketal nanoparticles: A new pH-sensitive biodegradable drug delivery vehicle. Bioconj. Chem. 2005, 16, 1340–1342. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Hong, C.Y.; Pan, C.Y. Doxorubicin-loaded aromatic imine-contained amphiphilic branched star polymer micelles: Synthesis, self-assembly, and drug delivery. Int. J. Nanomed. 2015, 10, 3623–3640. [Google Scholar]
- Liu, J.; Huang, Y.; Kumar, A.; Tan, A.; Jin, S.; Mozhi, A.; Liang, X.J. pH-sensitive nano-systems for drug delivery in cancer therapy. Biotechnol. Adv. 2014, 32, 693–710. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, G.; Tiwari, R.; Sriwastawa, B.; Bhati, L.; Pandey, S.; Pandey, P.; Bannerjee, S.K. Drug delivery systems: An updated review. Int. J. Pharm. Investig. 2012, 2, 2–11. [Google Scholar] [CrossRef] [PubMed]









| Code | In Feed (mol %) | In Copolymer (mol %) a | Mw b | PDI b | ||||
|---|---|---|---|---|---|---|---|---|
| H | D | B | H | D | B | |||
| BD | 0 | 16% | 8% | 0 | 11.9% | 3.6% | 1510 | 3.53 |
| HBD | 16% | 16% | 8% | 5.3% | 14.1% | 2.9% | 850 | 1.55 |
| Group | Copolymer (mg) | IMQ (mg) | Size (nm) | PDI | Zeta (mv) | E.E. a (%) |
|---|---|---|---|---|---|---|
| BD1 | 1 | 5 | 230 ± 12 | 0.22 ± 0.04 | −27.9 ± 2.0 | 1.23 ± 212 |
| BD5 | 5 | 5 | 163 ± 19 | 0.22 ± 0.01 | −28.1 ± 2.4 | 1.29 ± 239 |
| HBD1 | 1 | 5 | 177 ± 8 | 0.19 ± 0.02 | −27.2 ± 2.3 | 0.66 ± 633 |
| HBD5 | 5 | 5 | 138 ± 6 | 0.20 ± 0.01 | −27.2 ± 27. | 1.40 ± 466 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, Y.-H.; Lee, T.-Y.; Fu, P.-C.; Lo, C.-L.; Chiang, Y.-T. Multifunctional Polymer Nanoparticles for Dual Drug Release and Cancer Cell Targeting. Polymers 2017, 9, 213. https://doi.org/10.3390/polym9060213
Wen Y-H, Lee T-Y, Fu P-C, Lo C-L, Chiang Y-T. Multifunctional Polymer Nanoparticles for Dual Drug Release and Cancer Cell Targeting. Polymers. 2017; 9(6):213. https://doi.org/10.3390/polym9060213
Chicago/Turabian StyleWen, Yu-Han, Tsung-Ying Lee, Ping-Chuan Fu, Chun-Liang Lo, and Yi-Ting Chiang. 2017. "Multifunctional Polymer Nanoparticles for Dual Drug Release and Cancer Cell Targeting" Polymers 9, no. 6: 213. https://doi.org/10.3390/polym9060213
APA StyleWen, Y. -H., Lee, T. -Y., Fu, P. -C., Lo, C. -L., & Chiang, Y. -T. (2017). Multifunctional Polymer Nanoparticles for Dual Drug Release and Cancer Cell Targeting. Polymers, 9(6), 213. https://doi.org/10.3390/polym9060213