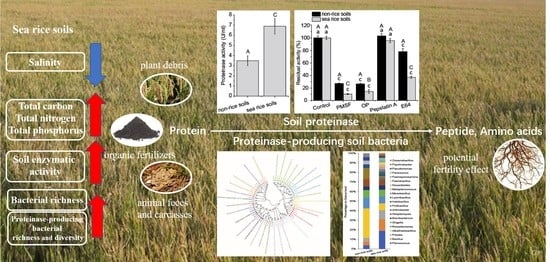
Impact of Sea Rice Planting on Enzymatic Activity and Microbial Community of Coastal Soils: Focus on Proteinase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling of Sea Rice Soils
2.2. Detection of Soil Physicochemical Characteristics and Proteinase Activity
2.3. DNA Amplification, Sequencing, and Analysis
2.4. Colony Count and Isolation of Culturable Proteinase-Producing Bacteria
2.5. 16S rDNA Sequencing and Phylogenetic Analysis
2.6. Proteinase Production Ability Detection of the Isolated Strains
2.7. Inhibitor Assay on Bacterial Extracellular Proteinase
2.8. Statistical Analysis
3. Results
3.1. Physicochemical Characteristics and Proteinase Activity of the Soil Samples
3.2. Alpha Diversity and Taxonomy Composition Analysis of Bacterial Community
3.3. Diversity Analysis of Proteinase-Producing Bacteria in Coastal Rice Soils
3.4. Diversity Analysis of the Bacterial Extracellular Proteinases
4. Discussion
4.1. The Impact of Sea Rice Planting on Physicochemical and Enzymatic Properties of Coastal Soils
4.2. The Impact of Sea Rice Planting on the Microbial Richness and Diversity of Coastal Soils
4.3. The Dominant Bacterial Community and Proteinase-Producing Bacteria in Coastal Rice Soils
4.4. The Potential Application of the Soil Proteinase and Bacterial Extracellular Proteinase in the Rice Planting
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Karstens, S.; Buczko, U.; Jurasinski, G.; Peticzka, R.; Glatzel, S. Impact of adjacent land use on coastal wetland sediments. Sci. Total Environ. 2016, 550, 337–348. [Google Scholar] [CrossRef]
- Long, X.H.; Liu, L.P.; Shao, T.Y.; Shao, H.B.; Liu, Z.P. Developing and sustainably utilize the coastal mudflat areas in China. Sci. Total Environ. 2016, 569–570, 1077–1086. [Google Scholar] [CrossRef]
- Gao, J.G.; Zhu, X.G. The legacies of the “Father of Hybrid Rice” and the seven representative achievements of Chinese rice research: A pioneering perspective towards sustainable development. Front. Plant Sci. 2023, 14, 1087768. [Google Scholar] [CrossRef]
- Wang, P.; Liu, Y.; Li, L.; Cheng, K.; Zheng, J.; Zhang, X.; Zheng, J.; Joseph, S.; Pan, G. Long-term rice cultivation stabilizes soil organic carbon and promotes soil microbial activity in a salt marsh derived soil chronosequence. Sci. Rep. 2015, 5, 15704. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, Q.; Chen, Y.; Dai, Q.; Hu, J. Dynamic change in enzyme activity and bacterial community with long-term rice cultivation in mudflats. Curr. Microbiol. 2019, 76, 361–369. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Greenfield, L.M.; Hill, P.W.; Paterson, E.; Baggs, E.M.; Jones, D.L. Do plants use root-derived proteases to promote the uptake of soil organic nitrogen? Plant Soil 2020, 456, 355–367. [Google Scholar] [CrossRef]
- Pathan, S.I.; Ceccherini, M.T.; Hansen, M.A.; Giagnoni, L.; Ascher, J.; Arenella, M.; Sørensen, S.J.; Pietramellara, G.; Nannipieri, P.; Renella, G. Maize lines with different nitrogen use efficiency select bacterial communities with different β-glucosidase-encoding genes and glucosidase activity in the rhizosphere. Biol. Fertil. Soils 2015, 51, 995–1004. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.-X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef]
- Staley, C.; Breuillin-Sessoms, F.; Wang, P.; Kaiser, T.; Venterea, R.T.; Sadowsky, M.J. Urea amendment decreases microbial diversity and selects for specific nitrifying strains in eight contrasting agricultural soils. Front. Microbiol. 2018, 9, 634. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Zheng, Y.; Hou, L.; Gao, D.; Chen, F.; Pei, C.; Dong, H.; Liang, X.; Liu, M. Microbial dynamics and activity of denitrifying anaerobic methane oxidizers in China’s estuarine and coastal wetlands. Sci. Total Environ. 2021, 806, 150425. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Guo, L.; Sun, J.; Wang, Y.; She, Z.; Gao, M.; Zhao, Y. Accelerating waste sludge hydrolysis with alkyl polyglucose pretreatment coupled with biological process of thermophilic bacteria: Hydrolytic enzyme activity and organic matters transformation. J. Environ. Manag. 2019, 247, 161–168. [Google Scholar] [CrossRef] [PubMed]
- López-Aizpún, M.; Arango-Mora, C.; Santamaría, C.; Lasheras, E.; Santamaría, J.M.; Ciganda, V.S.; Cárdenas, L.M.; Elustondo, D. Atmospheric ammonia concentration modulates soil enzyme and microbial activity in an oak forest affecting soil microbial biomass. Soil Biol. Biochem. 2018, 116, 378–387. [Google Scholar] [CrossRef]
- Li, H.; Chi, Z.; Li, J.; Wu, H.; Yan, B. Bacterial community structure and function in soils from tidal freshwater wetlands in a Chinese delta: Potential impacts of salinity and nutrient. Sci. Total Environ. 2019, 696, 134029. [Google Scholar] [CrossRef]
- Lemanowicz, J.; Haddad, S.A.; Bartkowiak, A.; Lamparski, R.; Wojewódzki, P. The role of an urban park’s tree stand in shaping the enzymatic activity, glomalin content and physicochemical properties of soil. Sci. Total Environ. 2020, 741, 140446. [Google Scholar] [CrossRef]
- Li, Y.; Wu, C.; Zhou, M.; Wang, E.T.; Zhang, Z.; Liu, W.; Ning, J.; Xie, Z. Diversity of cultivable protease-producing bacteria in Laizhou Bay sediments, Bohai Sea, China. Front. Microbiol. 2017, 8, 405. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.Y.; Han, X.X.; Chen, X.L.; Dang, H.Y.; Xie, B.B.; Qin, Q.L.; Shi, M.; Zhou, B.C.; Zhang, Y.Z. Diversity of cultivable protease-producing bacteria in sediments of Jiaozhou Bay, China. Front. Microbiol. 2015, 6, 1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Liu, W.; Xing, S.; Zhang, X.; He, H.; Chen, J.; Bielicki, J.K.; Zhou, M. Diversity of protease-producing bacteria in the soils of the South Shetland Islands, Antarctica. Antonie Leeuwenhoek 2021, 114, 457–464. [Google Scholar] [CrossRef]
- Butt, M.Q.; Zeeshan, N.; Ashraf, N.M.; Akhtar, M.A.; Ashraf, H.; Afroz, A.; Shaheen, A.; Naz, S. Environmental impact and diversity of protease-producing bacteria in areas of leather tannery effluents of Sialkot, Pakistan. Environ. Sci. Pollut. Res. Int. 2021, 28, 54842–54851. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Bu, J.; Long, H.; Zhang, X.; Cai, X.; Huang, A.; Ren, W.; Xie, Z. Community structure of protease-producing bacteria cultivated from aquaculture systems: Potential impact of a tropical environment. Front Microbiol. 2021, 12, 638129. [Google Scholar] [CrossRef]
- Choi, G.M.; Im, W.T. Paraburkholderia azotifigens sp. nov., a nitrogen-fixing bacterium isolated from paddy soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 310–316. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, S.Y.; Whang, K.S. Cellulomonas citrea sp. nov., isolated from paddy soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 5304–5311. [Google Scholar] [CrossRef] [PubMed]
- Moghadam, S.V.; Jafarzadeh, A.; Vadde, K.K.; Matta, A.; Dessouky, S.; Hutchinson, J. Composition of soil bacterial communities associated with urban stormwater detention basins and their predicted functional roles in N cycle. J. Appl. Microbiol. 2023, 134, lxad163. [Google Scholar] [CrossRef]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, G.; Guo, X.; Li, Y.; Ji, N.; Xu, X.; Sun, Q.; Yang, J. Diversity of the protease-producing bacteria and their extracellular protease in the coastal mudflat of Jiaozhou Bay, China: In response to clam naturally growing and aquaculture. Front. Microbiol. 2023, 14, 1164937. [Google Scholar] [CrossRef] [PubMed]
- Afoshin, A.; Tishchenko, S.; Gabdulkhakov, A.; Kudryakova, I.; Galemina, I.; Zelenov, D.; Leontyevskaya, E.; Saharova, S.; Leontyevskaya Vasilyeva, N. Structural and functional characterization of β-lytic protease from Lysobacter capsici VKM B-2533(T). Int. J. Mol. Sci. 2022, 23, 16100. [Google Scholar] [CrossRef] [PubMed]
- Sumathi, C.; Dillibabu, V.; Madhuri, D.K.; Priya, D.M.; Nagalakshmi, C.; Sekaran, G. Dietary inclusion of protease producing novel Pontibacter spp. and Bacillus megaterium as a probiotic enhances immune responses in Labeo rohita. Pak. J. Biol. Sci. 2014, 17, 451–461. [Google Scholar] [CrossRef] [Green Version]
- Solihin, J.; Waturangi, D.E.; Purwadaria, T. Induction of amylase and protease as antibiofilm agents by starch, casein, and yeast extract in Arthrobacter sp. CW01. BMC Microbiol. 2021, 21, 232. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, R.; Caballero, A.; Tang, A.; Bierdeman, M. Pseudomonas aeruginosa keratitis: Protease IV and PASP as corneal virulence mediators. Microorganisms 2019, 7, 281. [Google Scholar] [CrossRef] [Green Version]
- Miao, H.; Zhe, Y.; Xiang, X.; Cao, Y.; Han, N.; Wu, Q.; Huang, Z. Enhanced extracellular expression of a Ca(2+)- and Mg(2+)- dependent hyperthermostable protease EA1 in Bacillus subtilis via systematic screening of optimal signal peptides. J. Agric. Food Chem. 2022, 70, 15830–15839. [Google Scholar] [CrossRef]
- Xin, Y.; Sun, Z.; Chen, Q.; Wang, J.; Wang, Y.; Luogong, L.; Li, S.; Dong, W.; Cui, Z.; Huang, Y. Purification and characterization of a novel extracellular thermostable alkaline protease from Streptomyces sp. M30. J. Microbiol. Biotechnol. 2015, 25, 1944–1953. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ding, N.F.; Fu, Q.L.; Brookes, P.C.; Xu, J.M.; Guo, B.L.; Lin, Y.C.; Li, H.; Li, N.Y. The influence of soil properties on the size and structure of bacterial and fungal communities along a paddy soil chronosequence. Eur. J. Soil Biol. 2016, 76, 9–18. [Google Scholar] [CrossRef]
- Canfora, L.; Bacci, G.; Pinzari, F.; Lo Papa, G.; Dazzi, C.; Benedetti, A. Salinity and bacterial diversity: To what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS ONE 2014, 9, e106662. [Google Scholar] [CrossRef] [Green Version]
- Van Horn, D.J.; Okie, J.G.; Buelow, H.N.; Gooseff, M.N.; Barrett, J.E.; Takacs-Vesbach, C.D. Soil microbial responses to increased moisture and organic resources along a salinity gradient in a polar desert. Appl. Environ. Microbiol. 2014, 80, 3034–3043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Cui, Y.; Chen, Y.; Lin, X.; Huang, H.; Bao, S. Diversity of Bacillus-like bacterial community in the sediments of the Bamenwan mangrove wetland in Hainan, China. Can. J. Microbiol. 2017, 63, 238–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Hu, H.; Liu, Y.R.; Xiao, K.Q.; Cheng, F.S.; Li, J.; Xiao, T. Bacterial composition and spatiotemporal variation in sediments of Jiaozhou Bay, China. J. Soils Sediments 2014, 15, 1–13. [Google Scholar] [CrossRef]
- Hou, Y.; Zeng, W.; Ao, C.; Luo, Y.; Wang, Z.; Hou, M.; Huang, J. Bacillus atrophaeus WZYH01 and Planococcus soli WZYH02 Improve Salt Tolerance of Maize (Zea mays L.) in Saline Soil. Front. Plant Sci. 2022, 13, 891372. [Google Scholar] [CrossRef]
- Sinimol, S.; Sarika, A.R.; Jayakumaran Nair, A. Diversity and antagonistic potential of marine microbes collected from south-west coast of India. 3 Biotech 2015, 6, 7. [Google Scholar] [CrossRef] [Green Version]
- Farha, A.K.; Tr, T.; Purushothaman, A.; Salam, J.A.; Hatha, A.M. Phylogenetic diversity and biotechnological potentials of marine bacteria from continental slope of eastern Arabian Sea. J. Genet. Eng. Biotechnol. 2018, 16, 253–258. [Google Scholar] [CrossRef]
- Wagh, V.S.; Ram, H.; Dastager, S.G. Priestia veravalensis sp. nov., isolated from coastal sample. Arch. Microbiol. 2021, 203, 4839–4845. [Google Scholar] [CrossRef]
- Krishna, P.S.; Raghunathan, S.; Prakash, J.S.S. Comparative genome analysis of Alkalihalobacillus okhensis Kh10-101 T reveals insights into adaptive mechanisms for halo-alkali tolerance. 3 Biotech 2021, 11, 392. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Torre, S.; Carro, L.; Igual, J.M.; Montero-Calasanz, M.D.C. Rossellomorea arthrocnemi sp. nov., a novel plant growth-promoting bacterium used in heavy metal polluted soils as a phytoremediation tool. Int. J. Syst. Evol. Microbiol. 2021, 71, 005015. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; You, X.; Tang, Z.; Zhu, T.; Liu, B.; Chen, M.-X.; Xu, Y.; Liu, T.-Y. Isolation and identification of plant growth-promoting rhizobacteria from tall fescue rhizosphere and their functions under salt stress. Physiol. Plant. 2022, 174, e13817. [Google Scholar] [CrossRef] [PubMed]
- Desale, P.; Patel, B.; Singh, S.; Malhotra, A.; Nawani, N. Plant growth promoting properties of Halobacillus sp. and Halomonas sp. in presence of salinity and heavy metals. J. Basic Microbiol. 2014, 54, 781–791. [Google Scholar] [CrossRef]
- Phazna, T.A.; Ngashangva, N.; Yentrembam, R.B.S.; Maurya, R.; Mukherjee, P.; Sharma, C.; Verma, P.K.; Sarangthem, I. Draft genome sequence and functional analysis of Lysinibacillus xylanilyticus t26, a plant growth-promoting bacterium isolated from Capsicum chinense rhizosphere. J. Biosci. 2022, 47, 36. [Google Scholar] [CrossRef]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.C. Current knowledge and perspectives of Paenibacillus: A review. Microb. Cell Factories 2016, 15, 203. [Google Scholar] [CrossRef] [Green Version]
- Davies, B.; Coulter, J.A.; Pagliari, P.H. Soil enzyme activity behavior after urea nitrogen application. Plants 2022, 11, 2247. [Google Scholar] [CrossRef]
- Sofo, A.; Khanghahi, M.Y.; Curci, M.; Reyes, F.; Briones, M.J.I.; Sarneel, J.M. Earthworm-Driven Changes in Soil Chemico-Physical Properties, Soil Bacterial Microbiota, Tree/Tea Litter Decomposition, and Plant Growth in a Mesocosm Experiment with Two Plant Species. Plants 2023, 12, 1216. [Google Scholar] [CrossRef]
- Moreau, D.; Richard, D.B.; Roger, D.F.; David, L.J.; Laurent, P. A plant perspective on nitrogen cycling in the rhizosphere. Funct. Ecol. 2019, 33, 540–552. [Google Scholar] [CrossRef] [Green Version]
- Farzadfar, S.; Knight, J.D.; Congreves, K.A. Soil organic nitrogen: An overlooked but potentially significant contribution to crop nutrition. Plant Soil 2021, 462, 7–23. [Google Scholar] [CrossRef]
- Kawade, K.; Tabeta, H.; Ferjani, A.; Hirai, M.Y. The Roles of Functional Amino Acids in Plant Growth and Development. Plant Cell Physiol. 2023, pcad071. [Google Scholar] [CrossRef]
- Cai, S.; Zhang, Y.; Hu, A.; Liu, M.; Wu, H.; Wang, D.; Zhang, W. Dissolved organic matter transformation mechanisms and process optimization of wastewater sludge hydrothermal humification treatment for producing plant biostimulants. Water Res. 2023, 235, 119910. [Google Scholar] [CrossRef] [PubMed]
- Kamal, S.; Rehman, S.; Iqbal, H. Biotechnological valorization of proteases: From hyper-production to industrial exploitation–A review. Environ. Prog. Sustain. Energy 2016, 36, 511–522. [Google Scholar] [CrossRef]
- Jiao, S.; Chen, W.; Wang, J.; Du, N.; Li, Q.; Wei, G. Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome 2018, 6, 146. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.Y.; Wang, G.L.; Li, D.; Zhao, D.L.; Qin, Q.L.; Chen, X.L.; Chen, B.; Zhou, B.C.; Zhang, X.Y.; Zhang, Y.Z. Diversity of both the cultivable protease-producing bacteria and bacterial extracellular proteases in the coastal sediments of King George Island, Antarctica. PLoS ONE 2013, 8, e79668. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Station | Location | Temperature | pH | Salinity | TC | TN | TP |
|---|---|---|---|---|---|---|---|
| (E, N) | (°C) | (g/kg) | (g/kg) | (g/kg) | (g/kg) | ||
| sea rice field | 120°11′41″, 36°19′10″ | 16.8 | 7.98 ± 0.05 | 2.15 ± 0.1 * | 17.48 ± 0.62 ** | 0.92 ± 0.06 * | 0.53 ± 0.07 * |
| non-rice region | 120°11′29″, 36°18′56″ | 16.8 | 8.17 ± 0.06 | 2.6 ± 0.18 | 14.75 ± 0.34 | 0.68 ± 0.06 | 0.35 ± 0.02 |
| Sample | Coverage (%) | Diversity Index | Richness Index | |||
|---|---|---|---|---|---|---|
| Shannon | Simpson × 10−3 | Sobs | Ace | Chao | ||
| sea rice soils | 98.05 ± 0.24 | 5.85 ± 0.46 | 13.90 ± 10.69 | 1789 ± 110 * | 2191 ± 119 * | 2183 ± 142 * |
| reference soils | 98.52 ± 0.12 | 5.88 ± 0.21 | 7.18 ± 2.49 | 1515 ± 70 | 1800 ± 34 | 1733 ± 31 |
| Genera | Strains | Inhibition Ratio (%) 1 | |||
|---|---|---|---|---|---|
| PMSF 2 | OP 3 | Pepstatin A 4 | E64 5 | ||
| Alkalihalobacillus | G2-7a | 39.11 | 45.12 | 27.77 | 10.54 |
| E2-1 | 12.36 | 30.39 | — | — | |
| E3-3 | 17.30 | 56.89 | — | — | |
| Bacillus | G1-7 | 40.87 | 45.77 | 20.69 | 6.44 |
| E2-6 | 46.86 | — | — | — | |
| E1-2 | 24.86 | 26.83 | — | — | |
| E3-15 | 27.57 | 15.49 | 6.05 | — | |
| E1-9 | 48.08 | 4.24 | — | — | |
| E1-8 | 38.56 | 56.64 | 2.59 | 21.82 | |
| Fictibacillus | G1-8 | 53.62 | — | — | — |
| G1-13 | 34.23 | 14.92 | 11.53 | 4.29 | |
| E2-8 | 47.89 | 13.71 | — | — | |
| Lysinibacillus | E1-4 | 34.67 | — | — | — |
| Priestia | G1-17 | 11.77 | 65.16 | — | — |
| E3-12 | 14.74 | 41.86 | — | — | |
| E1-21 | 40.21 | 50.69 | 18.08 | 6.75 | |
| E1-7 | 40.65 | 42.92 | — | — | |
| Rossellomorea | G2-4 | 41.98 | 8.21 | — | — |
| G1-6 | 20.82 | 6.27 | — | — | |
| E1-1b | 14.17 | 28.94 | — | — | |
| E1-13 | 16.21 | 5.88 | — | — | |
| Metaplanococcus | E1-18 | 52.16 | 21.12 | 13.44 | 9.29 |
| Planococcus | G1-5 | 53.81 | 17.92 | — | — |
| G1-15 | 34.92 | 39.25 | — | — | |
| E1-23 | 39.37 | — | — | — | |
| E1-14 | 29.86 | — | — | — | |
| E2-10 | 58.30 | 28.81 | — | — | |
| E1-5 | 21.21 | 20.06 | — | — | |
| Psychrobacter | E3-9 | 43.66 | 31.06 | 19.00 | 10.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Liu, Z.; Zhang, M.; Zhu, X.; Wang, M.; Xu, X.; Liu, G. Impact of Sea Rice Planting on Enzymatic Activity and Microbial Community of Coastal Soils: Focus on Proteinase. Agronomy 2023, 13, 2089. https://doi.org/10.3390/agronomy13082089
Yang J, Liu Z, Zhang M, Zhu X, Wang M, Xu X, Liu G. Impact of Sea Rice Planting on Enzymatic Activity and Microbial Community of Coastal Soils: Focus on Proteinase. Agronomy. 2023; 13(8):2089. https://doi.org/10.3390/agronomy13082089
Chicago/Turabian StyleYang, Jie, Zhiyun Liu, Mingyi Zhang, Xiaolong Zhu, Mingyi Wang, Xingfeng Xu, and Guangchao Liu. 2023. "Impact of Sea Rice Planting on Enzymatic Activity and Microbial Community of Coastal Soils: Focus on Proteinase" Agronomy 13, no. 8: 2089. https://doi.org/10.3390/agronomy13082089
APA StyleYang, J., Liu, Z., Zhang, M., Zhu, X., Wang, M., Xu, X., & Liu, G. (2023). Impact of Sea Rice Planting on Enzymatic Activity and Microbial Community of Coastal Soils: Focus on Proteinase. Agronomy, 13(8), 2089. https://doi.org/10.3390/agronomy13082089