Biological Exploration and Physicochemical Characteristics of Tomato Brown Rugose Fruit Virus in Several Host Crops
Abstract
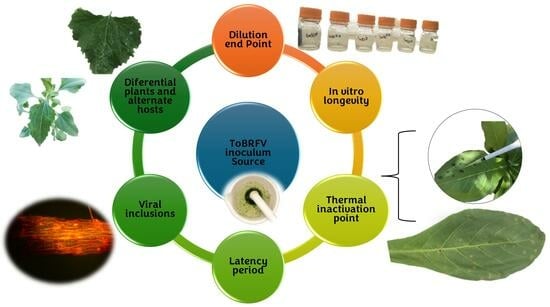
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Condition
2.2. Dilution Endpoint from Inoculations on Tomato Plants var. 172–300
2.2.1. Evaluation of Agronomic Variables
2.2.2. Determination of Viral Concentration by DAS-ELISA
2.2.3. Pearson’s Correlation Analysis
2.3. Determination of Latency and Incubation Period from Inoculations in Tomato Plants var. 172–300
2.3.1. Determination of Viral Concentration by DAS-ELISA
2.3.2. Determination of Severity and Area under the Disease Progress Curve (AUDPC)
2.4. The Presence of ToBRFV Infective Sap in Plants of Nicotiana longiflora
2.4.1. Thermal Inactivation Points of ToBRFV in Plants of Nicotiana longiflora
2.4.2. In Vitro Longevity from ToBRFV Inoculations in Nicotiana longiflora Plants
2.4.3. In Vitro Longevity of ToBRFV in N. Longiflora Plants
2.4.4. Persistence of ToBRFV in Geometric Progression in Nicotiana longiflora Plants
2.5. Viral Inclusions in Tomato var. 172–300 and Nicotiana Tabacum Plants
2.6. Study of Alternate Hosts for ToBRFV Identified in the Natural Environment
2.6.1. Differential Plant Diagnosis
2.6.2. Potential Hosts of ToBRFV
2.6.3. Confirmation by DAS-ELISA
2.7. Statistical Analysis
3. Results
3.1. Confirmation of the ToBRFV
3.2. Dilution Endpoint from ToBRFV Inoculations in Tomato Plants
3.3. Determination of Latency and Incubation Period in Tomato Plants var. 172–300 Inoculated with ToBRFV
3.4. Presence of ToBRFV Infective Sap in Nicotiana longiflora Plants
3.4.1. Thermal Inactivation Points of ToBRFV in Nicotiana longiflora Plants
3.4.2. In Vitro Longevity of Nicotiana longiflora following Inoculation with ToBRFV
3.4.3. Persistence of ToBRFV in Geometric Progression in Nicotiana longiflora Plants
3.5. Viral Inclusions in Tomato var. 172–300 and Nicotiana Tabacum Plants in Response to ToBRFV Infection
3.6. Diagnosis of ToBRFV by Differential Plants
Host Plants Susceptible to Inoculation with ToBRFV
4. Discussion
4.1. Impact of ToBRFV Infection at Diverse Sap Concentrations in Tomato var. 172–300 Plants
4.2. Determination of the Latency and Incubation Period of ToBRFV in Tomato Plants var. 172–300
4.3. Presence of ToBRFV Infective Sap in Nicotiana longiflora Plants
4.4. Viral Inclusions
4.5. Diagnostic Hosts and Alternate Reservoirs of ToBRFV
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruíz, A.M.Y.; Montes, M.J.A.; Castañón, G.J.H.; Gutiérrez, M.F.A.; Hernández, G.M.; López, L.H.; Ruíz, V.V.M.; Villalobos, M.J.J. Uso de la harina del pez diablo (Pterygoplichthys spp.) en la fertilización orgánica del tomate (Solanum lycopersicum L.). Rev. Int. Contam. Ambient. 2023, 39, 159–169. [Google Scholar] [CrossRef]
- González-Concha, L.F.; Ramírez-Gil, J.G.; García-Estrada, R.S.; Rebollar-Alviter, Á.; Tovar-Pedraza, J.M. Spatiotemporal Analyses of Tomato Brown Rugose Fruit Virus in Commercial Tomato Greenhouses. Agronomy 2021, 11, 1268. [Google Scholar] [CrossRef]
- Salem, N.; Mansour, A.; Ciuffo, M.; Falk, B.W.; Turina, M. A new Tobamovirus infecting tomato crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar] [CrossRef]
- Luria, N.; Smith, E.; Reingold, V.; Bekelman, I.; Lapidot, M.; Levin, I. A new Israeli Tobamovirus isolate infects tomato plants harboring Tm-22 resistance genes. PLoS ONE 2017, 12, e0170429. [Google Scholar] [CrossRef] [PubMed]
- Cambrón-Crisantos, J.M.; Rodríguez-Mendoza, J.; Valencia-Luna, J.B.; Alcasio Rangel, S.; de Jesús García-Ávila, C.; López-Buenfil, J.A.; Ochoa-Martínez, D.L.; Cambrón-Crisantos, J.M.; Rodríguez-Mendoza, J.; Valencia-Luna, J.B.; et al. First Report of Tomato Brown Rugose Fruit Virus (ToBRFV) in Michoacan, Mexico. Rev. Mex. Fitopatol. 2019, 37, 185–192. [Google Scholar] [CrossRef]
- Camacho-Beltrán, E.; Pérez-Villarreal, A.; Leyva-López, N.E.; Rodríguez-Negrete, E.A.; Ceniceros-Ojeda, E.A.; Méndez-Lozano, J. Occurrence of Tomato Brown Rugose Fruit Virus Infecting Tomato Crops in Mexico. Plant Dis. 2019, 103, 1440. [Google Scholar] [CrossRef]
- Van de Vossenberg, B.T.; Visser, M.; Bruinsma, M.; Koenraadt, H.M.S.; Westenberg, M. Real-time tracking of Tomato Brown Rugose Fruit Virus (ToBRFV) outbreaks in the Netherlands using Nextstrain. PLoS ONE 2020, 15, e0234671. [Google Scholar] [CrossRef]
- EPPO. European and Mediterranean Plant Protection Organization: Tomato Brown Rugose Fruit Virus. Global Database Distribution World. Available online: https://gd.eppo.int/taxon/TOBRFV/distribution (accessed on 14 April 2023).
- Ortiz-Martínez, L.E.; Ochoa-Martínez, D.L. Elicitors and biostimulants in the production of tomato infected with Tomato Brown Rugose Fruit Virus. J. Plant Dis. Prot. 2023, 130, 351–360. [Google Scholar] [CrossRef]
- Iwanowski, D. Concerning the mosaic disease of the tobacco plant. In St. Petersb. Acad. Imp. Sci. Bull., Phytopathological Classics Number 7 (1942); Translator Johnson, J.; American Phytopathological Society Press: St. Paul, MN, USA, 1892; Volume 35, pp. 67–70. [Google Scholar]
- Dombrovsky, A.; Tran-Nguyen, L.T.T.; Jones, R.A.C. Cucumber Green Mottle Mosaic Virus: Rapidly increasing global distribution, etiology, epidemiology, and management. Annu. Rev. Phytopathol. 2017, 55, 231–256. [Google Scholar] [CrossRef]
- Khamphirapaeng, P.; Cheewangkoon, R.; McGovern, R.J.; Wong, S.M.; To-Anun, C. Detection of Tobacco Mosaic Virus in petunia and tobacco by light microscopy using a simplified inclusion body staining technique. Int. J. Agric. Technol. 2017, 13, 163–168. Available online: http://www.ijat-aatsea.com/ (accessed on 28 December 2023).
- Jewehan, A.; Kiemo, F.W.; Salem, N. Isolation and molecular characterization of a Tomato Brown Rugose Fruit Virus mutant breaking the tobamovirus resistance found in wild Solanum species. Arch Virol. 2022, 167, 1559–1563. [Google Scholar] [CrossRef] [PubMed]
- Chanda, B.; Gilliard, A.; Jaiswal, N.; Ling, K.S. Comparative analysis of host range, ability to infect tomato cultivars with Tm-22 gene, and real-time reverse transcription PCR detection of Tomato Brown Rugose Fruit Virus. Plant Dis. 2021, 105, 3643–3652. [Google Scholar] [CrossRef] [PubMed]
- Chanda, B.; Rivera, Y.; Nunziata, S.O.; Galvez, M.E.; Gilliard, A.; Ling, K.S. Complete genome sequence of a Tomato Brown Rugose Fruit Virus isolated in the United States. Microbiol. Resour. Announc. 2020, 9, e00630-20. [Google Scholar] [CrossRef]
- Vaisman, M.; Hak, H.; Arazi, T.; Spiegelman, Z. The impact of tobamovirus infection on root development involves induction of auxin response factor 10a in tomato. Plant Cell Physiol. 2022, 63, 1980–1993. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, J.; Nourinejhad Zarghani, S.; Kroschewski, B.; Büttner, C.; Bandte, M. Cleaning of Tomato Brown Rugose Fruit Virus (ToBRFV) from Contaminated Clothing of Greenhouse Employees. Horticulturae 2022, 8, 751. [Google Scholar] [CrossRef]
- Eads, A.; Groth-Helms, D.; Davenport, B.; Cha, X.; Li, R.; Walsh, C.; Schuetz, K. The commercial validation of three Tomato brown rugose fruit virus assays. PhytoFrontiers 2023, 3, 206–213. [Google Scholar] [CrossRef]
- Rodríguez-Mendoza, J.; García-Ávila, C.J.; López-Buenfil, J.A.; Araujo-Ruiz, K.; Quezada-Salinas, A.; Cambrón-Crisantos, J.M.; Ochoa-Martínez, D.L. Identificación de Tomato Brown Rugose Fruit Virus por RT-PCR de una región codificante de la replicasa (RdRP). Rev. Mex. Fitopatol. 2019, 37, 345–356. [Google Scholar] [CrossRef]
- Vasquez-Gutierrez, U.; Delgado-Ortiz, J.C.; Frías-Treviño, G.A.; Aguirre-Uribe, L.A.; Olivas, A.F. Pathogenicity of three Tomato brown rugose fruit virus isolates and response in tomato (Solanum lycopersicum L.). In Proceedings of the 3rd International Electronic Conference on Agronomy, Basel, Switzerland, 15–30 October 2023. [Google Scholar]
- Steiner, A.A. A universal method for preparing nutrient solutions of a certain desired composition. Plant Soil 1961, 15, 134–154. [Google Scholar] [CrossRef]
- Vasquez-Gutierrez, U.; Frias-Treviño, G.A.; Delgado-Ortiz, J.C.; Aguirre-Uribe, L.A.; Flores-Olivas, A. Severity of Tomato Brown Rugose Fruit Virus in tomato (Solanum lycopersicum L.) from a region of Coahuila, México. Int. J. Hortic. Agric. Food Sci. 2023, 7, 1–6. [Google Scholar] [CrossRef]
- Fajinmi, A.A.; Dokunmu, A.O.; Akheituamen, D.O.; Onanuga, K.A. Incidence and infection rate of Maize streak virus disease by Cicadulina triangular on maize plants and its distribution from the lowest diseased leaf under tropical conditions. Arch. Phytopathol. Plant Prot. 2012, 45, 1591–1598. [Google Scholar] [CrossRef]
- Kone, N.; Asare-Bediako, E.; Silue, S.; Kone, D.; Koita, O.; Menzel, W.; Winter, S. Influence of planting date on incidence and severity of viral disease on cucurbits under field condition. Ann. Agric. Sci. 2017, 62, 99–104. [Google Scholar] [CrossRef]
- Jewehan, A.; Salem, N.; Tóth, Z. Evaluation of responses to Tomato Brown Rugose Fruit Virus (ToBRFV) and selection of resistant lines in Solanum habrochaites and Solanum peruvianum germplasm. J. Gen. Plant Pathol. 2022, 88, 187–196. [Google Scholar] [CrossRef]
- Ortiz-Martínez, L.E.; Ochoa-Martínez, D.L.; Rojas-Martínez, R.I.; Aranda-Ocampo, S.; Cruz, M.A.G. Respuesta de variedades de chile a la infección con Tomato Brown Rugose Fruit Virus. Summa Phytopathol. 2022, 47, 209–215. [Google Scholar] [CrossRef]
- Shaner, G. The Effect of Nitrogen Fertilization on the Expression of Slow-Mildewing Resistance in Knox Wheat. Phytopathology 1977, 67, 1051–1056. [Google Scholar] [CrossRef]
- Vargas-Mejía, P.; Rodríguez-Gómez, G.; Salas-Aranda, D. A Identificación y manejo del virus del fruto rugoso marrón del tomate en invernaderos en México. Arch. Virol. 2023, 168, 135. [Google Scholar] [CrossRef]
- Nourinejhad Zarghani, S.; Monavari, M.; Ehlers, J.; Hamacher, J.; Büttner, C.; Bandte, M. Comparison of Models for Quantification of Tomato Brown Rugose Fruit Virus Based on a Bioassay Using a Local Lesion Host. Plants 2022, 11, 3443. [Google Scholar] [CrossRef]
- Francki, R.I. Limited value of the thermal inactivation point, longevity in vitro and dilution endpoint as criteria for the characterization, identification, and classification of plant viruses. Intervirology 1980, 13, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Cárdena, S.E. Diagnóstico De Virus Mediante Inclusiones Virales, Microscopía Electrónica y Rango De Hospedantes, 3rd ed.; Colegio de Postgraduados, Instituto de Fitosanidad: Montecillo, Mexico, 1999; pp. 154–196. [Google Scholar]
- Salem, N.M.; Abumuslem, M.; Turina, M.; Samarah, N.; Sulaiman, A.; Abu-Irmaileh, B.; Ata, Y. New Weed Hosts for Tomato Brown Rugose Fruit Virus in Wild Mediterranean Vegetation. Plants 2022, 11, 2287. [Google Scholar] [CrossRef] [PubMed]
- Matzrafi, M.; Abu-Nassar, J.; Klap, C.; Smith, E.; Dombrovsky, A. Solanum elaeagnifolium and S. rostratum as potential hosts of the tomato brown rugose fruit virus. PLoS ONE 2022, 18, e0282441. [Google Scholar] [CrossRef]
- Sabra, A.; Amer, M.A.; Hussain, K.; Zakri, A.; AlShahwan, I.M.; Al-Saleh, M.A. Occurrence and Distribution of Tomato Brown Rugose Fruit Virus Infecting Tomato Crop in Saudi Arabia. Plants 2023, 11, 3157. [Google Scholar] [CrossRef]
- Zhang, S.; Griffiths, J.S.; Marchand, G.; Bernards, M.A.; Wang, A. Tomato Brown Rugose Fruit Virus: An emerging and rapidly spreading plant RNA virus that threatens tomato production worldwide. Mol. Plant Pathol. 2022, 23, 1262–1277. [Google Scholar] [CrossRef]
- Smith, E.; Dombrovsky, A. Aspects in Tobamovirus management in intensive agriculture. In Plant Diseases-Current Threats and Management Trends; IntechOpen: London, UK, 2019. [Google Scholar] [CrossRef]
- Jablonski, M.; Poghossian, A.; Keusgen, M. Detection of plant virus particles with a capacitive field-effect sensor. Anal. Bioanal. Chem. 2021, 413, 5669–5678. [Google Scholar] [CrossRef]
- Koh, S.H.; Li, H.; Sivasithamparam, K.; Admiraal, R.; Jones, M.G.K.; Wylie, S.J. Low root-to-root transmission of a tobamovirus, yellow tailflower mild mottle virus, and resilience of its virions. Plant Pathol. 2018, 67, 651–659. [Google Scholar] [CrossRef]
- Takács, A.; Kazinczi, G.; Pribék, D. Biological Decline of Solanum nigrum L. Due to Tobacco Mosaic Tobamovirus (TMV) Infection I. Growth and Nutrient Uptake. Acta Phytopathol. Entomol. Hung. 2001, 36, 9–14. [Google Scholar] [CrossRef]
- Bhat, A.I.; Rao, G.P. Symptoms of Virus-Infected Plants. In Characterization of Plant Viruses; Bhat, A.I., Rao, G.P., Eds.; Springer: New York, NY, USA, 2020; Volume 2, pp. 7–22. [Google Scholar]
- Van Mölken, T.; Stuefer, J.F. The potential of plant viruses to promote genotypic diversity via genotype× environment interactions. Ann. Bot. 2011, 107, 1391–1397. [Google Scholar] [CrossRef] [PubMed]
- Rys, M.; Juhász, C.; Surówka, E.; Janeczko, A.; Saja, D.; Tóbiás, I.; Gullner, G. Comparison of a compatible and an incompatible pepper-tobamovirus interaction by biochemical and non-invasive techniques: Chlorophyll a fluorescence, isothermal calorimetry, and FT-Raman spectroscopy. Plant Physiol. Biochem. 2014, 83, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, I.M.; Thomma, B.P. Cucumber Mosaic Virus: A Successful Pathogen That Rapidly Evolved from Emerging to Endemic in Tomato Crops. Mol. Plant Pathol. 2010, 11, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Kabas, A.; Fidan, H.; Kucukaydin, H.; Atan, H.N. Screening of wild tomato species and interspecific hybrids for resistance/tolerance to Tomato Brown Rugose Fruit Virus (ToBRFV). Chil. J. Agric. Res. 2022, 82, 189–196. [Google Scholar] [CrossRef]
- Mehle, N.; Bačnik, K.; Bajde, I.; Brodarič, J.; Fox, A.; Gutiérrez-Aguirre, I.; Vučurović, A. Tomato Brown Rugose Fruit Virus in aqueous environments–survival and significance of water-mediated transmission. Front. Plant Sci. 2023, 14, 1187920. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.Y.; Zhao, M.S.; Liu, L.Z.; Yang, G.L.; Geng, C.; Tian, Y.; Li, X.D. Biological and molecular characterization of Tomato Brown Rugose Fruit Virus and development of quadruplex RT-PCR detection. J. Integr. Agric. 2021, 20, 1871–1879. [Google Scholar] [CrossRef]
- Bernabé-Orts, J.M.; Torre, C.; Méndez-López, E.; Hernando, Y.; Aranda, M.A. Nuevos recursos para la detección específica y sensible del virus emergente del fruto rugoso marrón del tomate. Virus 2021, 13, 1680. [Google Scholar] [CrossRef]
- Pineda, M.; Olejníčková, J.; Cséfalvay, L.; Barón, M. Tracking viral movement in plants by means of chlorophyll fluorescence imaging. J. Plant Physiol. 2011, 168, 2035–2040. [Google Scholar] [CrossRef] [PubMed]
- Pineda, M.; Sajnani, C.; Barón, M. Changes induced by the Pepper mild mottle tobamovirus on the chloroplast proteome of Nicotiana benthamiana. Photosynth. Res 2010, 103, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Dombrovsky, A.; Mor, N.; Gantz, S.; Lachman, O.; Smith, E. Disinfection efficacy of Tobamovirus-contaminated soil in greenhouse-grown crops. Horticulturae 2022, 8, 563. [Google Scholar] [CrossRef]
- Schramm, G.; Engler, R. The latent period after infection with tobacco mosaic virus and virus nucleic acid. Nature 1958, 181, 916–917. [Google Scholar] [CrossRef]
- Yarwood, C.E. Latent Period and Generation Time for Two Plant Viruses. Am. J. Bot. 1952, 39, 613–618. [Google Scholar] [CrossRef]
- Broadbent, L. The epidemiology of tomato mosaic. XI. Seed transmission of TMV. Ann. Appl. Biol. 1965, 56, 177–205. [Google Scholar] [CrossRef]
- Reingold, V.; Lachman, O.; Belausov, E.; Koren, A.; Mor, N.; Dombrovsky, A. Epidemiological study of Cucumber Green Mottle Mosaic Virus in greenhouses enables reduction of disease damage in cucurbit production. Ann. Appl. Biol. 2016, 168, 29–40. [Google Scholar] [CrossRef]
- Li, R.; Zheng, Y.; Fei, Z.; Ling, K.S. First complete genome sequence of an emerging Cucumber Green Mottle Mosaic Virus isolate in North America. Genome Announc. 2015, 3, 10–1128. [Google Scholar] [CrossRef]
- Nash, D.; Ellmen, I.; Knapp, J.J.; Menon, R.; Overton, A.K.; Cheng, J.; Charles, T.C. A Novel Tiled-Amplicon Sequencing Assay Targeting the Tomato Brown Rugose Fruit Virus (ToBRFV) Genome Reveals Widespread Distribution in Municipal Wastewater Treatment Systems in the Province of Ontario, Canada. bioRxiv 2023, 11. [Google Scholar] [CrossRef]
- Lopez-Roblero, A.; Martínez Cano, D.J.; Diego-García, E.; Guillén-Navarro, G.K.; Iša, P.; Zarza, E. Metagenomic analysis of plant viruses in tropical fresh and wastewater. Environ. DNA 2023, 6, e416. [Google Scholar] [CrossRef]
- Bačnik, K.; Kutnjak, D.; Pecman, A.; Mehle, N.; Žnidarič, M.T.; Aguirre, I.G.; Ravnikar, M. Viromics and infectivity analysis reveal the release of infective plant viruses from wastewater into the environment. Water Res. 2020, 177, 115628. [Google Scholar] [CrossRef]
- Aguado-García, Y.; Taboada, B.; Morán, P.; Rivera-Gutiérrez, X.; Serrano-Vázquez, A.; Iša, P. Tobamoviruses can be frequently present in the oropharynx and gut of infants during their first year of life. Sci. Rep. 2020, 10, 13595. [Google Scholar] [CrossRef]
- Ehlers, J.; Nourinejhad Zarghani, S.; Liedtke, S.; Kroschewski, B.; Büttner, C.; Bandte, M. Análisis de la dispersión espacial del virus del fruto rugoso marrón del tomate en superficies en un sitio de producción comercial de tomate. Horticulturae 2023, 9, 611. [Google Scholar] [CrossRef]
- Samarah, N.; Sulaiman, A.; Salem, N.M. Disinfection treatments eliminated Tomato Brown Rugose Fruit Virus in tomato seeds. Eur. J. Plant Pathol. 2021, 159, 153–162. [Google Scholar] [CrossRef]
- Skelton, A.; Frew, L.; Ward, R.; Hodgson, R.; Forde, S.; McDonough, S.; Webster, G.; Chisnall, K.; Mynett, M.; Buxton-Kirk, A. Virus del fruto rugoso marrón del tomate: Eficacia de supervivencia y desinfección en superficies comunes de invernaderos. Virus 2023, 15, 2076. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, R.; Rohde, M.J.; Richert-Pöggeler, K.R.; Ziebell, H. To Be Seen or Not to Be Seen: Latent Infection by Tobamoviruses. Plants 2022, 11, 2166. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.D.; Finch, J.T.; Gibbs, A.J.; Hollings, M.; Shepherd, R.J.; Valenta, V.; Wetter, C. Sixteen groups of plant viruses. Virology 1971, 45, 356. [Google Scholar] [CrossRef]
- Edwardson, J.R.; Christie, R.G. Use of virus-induced inclusions in classification and diagnosis. Annu. Rev. Phytopathol. 1978, 16, 31. [Google Scholar] [CrossRef]
- Martelli, G.P.; Russo, M. Plant virus inclusion bodies. Adv. Virus Res. 1977, 21, 175. [Google Scholar] [CrossRef]
- Grangeon, R.; Jiang, J.; Wan, M.; Agbeci, H.; Zheng, J.F. Laliberte 6K2-induced vesicles can move cell to cell during Turnip mosaic virus infection. Front. Microbiol. 2013, 4, 351. [Google Scholar] [CrossRef] [PubMed]
- Maule, A.J.; Benitez-Alfonso, Y.; Faulkner, C. Plasmodesmata–membrane tunnels with attitude. Curr. Opin. Plant Biol. 2011, 14, 683–690. [Google Scholar] [CrossRef]
- Saito, T.; Yamanaka, K.; Okada, Y. Long-distance movement, and viral assembly of tobacco mosaic virus mutants. Virology 1990, 176, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Maule, A.J.; Palukaitis, P. Virus movement in infected plants. Crit. Rev. Plant Sci. 1991, 9, 457–473. [Google Scholar] [CrossRef]
- Hipper, C.; Brault, V.; Ziegler-Graff, V.; Revers, F. Viral and cellular factors involved in phloem transport of plant viruses. Front. Plant Sci. 2013, 4, 154. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.W.; Voinnet, O. Antiviral immunity directed by small RNAs. Cell 2007, 130, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Edwardson, J.R.; Christie, R.G.; Purcifull, D.E.; Petersen, M.A. Inclusions in diagnosing plant virus diseases. In Diagnosis of Plant Virus Diseases, 1st ed.; CRC Press: Boca Raton, FL, USA, 2019; pp. 101–128. [Google Scholar]
- Harries, P.A.; Schoelz, J.E.; Nelson, R.S. Intracellular transport of viruses and their components: Utilizing the cytoskeleton and membrane highways. Mol. Plant Microbe Interact. 2010, 23, 1381–1393. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Peña, E.J.; Amari, K.; Heinlein, M. Microtubules in viral replication and transport. Plant J. 2013, 75, 290–308. [Google Scholar] [CrossRef]
- Watanabe, Y.; Ohnishi, J.; Saitoh, H.; Hosokawa, D.; Okada, Y. Addition of nucleotides similar to deleted CAA repeats in the 5′ non-coding region of tomato mosaic virus RNA following propagation. J. Gen. Virol. 1996, 77, 2353–2357. [Google Scholar] [CrossRef]
- Roistacher, C.N. Graft-transmissible Diseases of Citrus. In Handbook for Detection and Diagnosis; IOCV, Riverside and FAO: Rome, Italy, 1991; p. 286. Available online: https://archive.org/details/bub_gb_9zY1uFGchAgC (accessed on 22 December 2023).
- Panno, S.; Caruso, A.G.; Barone, S.; Lo Bosco, G.; Rangel, E.A.; Davino, S. Spread of Tomato Brown Rugose Fruit Virus in Sicily and Evaluation of the Spatiotemporal Dispersion in Experimental Conditions. Agronomy 2020, 10, 834. [Google Scholar] [CrossRef]
- Izaguirre, M.M.; Mazza, C.A.; SvatoŠ, A.; Baldwin, I.T.; BallarÉ, C.L. La radiación solar ultravioleta B y la herbivoría de insectos desencadenan respuestas fenólicas parcialmente superpuestas en Nicotiana attenuata y Nicotiana longiflora. An. Botánica 2007, 99, 103–109. [Google Scholar] [CrossRef]
- Valleau, W.D.; Stokes, G.W.; Johnson, E.M. Nueve años de experiencia con el factor Nicotiana longiflora para resistencia a Phytophthora parasitica var. nicotianae en el control del jarrete negro. Tabaco 1960, 150, 12. [Google Scholar]
- Ahmad, B.; Jan, Q.; Bashir, S.; Nisar, M.; Shaheen, F.; Ahmad, M. Pharmacological and biological investigations of Chenopodium murale Linn. Asian J. Plant Sci. 2003, 2, 1107–1111. [Google Scholar] [CrossRef]
- Florentine, S.K.; Weller, S.; Graz, P.F.; Westbrooke, M.; Florentine, A.; Javaid, M.; Dowling, K. Influence of selected environmental factors on seed germination and seedling survival of the arid zone invasive species tobacco bush (Nicotiana glauca R. Graham). Rangel. J. 2016, 38, 417–425. [Google Scholar] [CrossRef]
- Solberg, R.A.; Bald, J.G. Distribution of a natural and an alien form of Tobacco Mosaic Virus in the shoot apex of Nicotiana glauca Grah. Virology 1963, 21, 300–308. [Google Scholar] [CrossRef]
- Wei, S.; Xu, L.; Dai, H. Ornamental hyperaccumulator Mirabilis jalapa L. phytoremediation combine contaminated soil enhanced by some chelators and surfactants. Environ. Sci. Pollut. Res. 2018, 25, 29699–29704. [Google Scholar] [CrossRef]










| Treatments | Agronomic Variables Evaluated | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PH (cm) | FFW (g) | FDW (g) | RFW (g) | RDW (g) | SD (mm) | Chlorophyll (SPAD) | N | Severity A | |
| Control | 35.4 ± 1.14 a | 13.34 ± 1.54 a | 6.94 ± 0.36 a | 7.33 ± 0.30 a | 4.46 ± 1.96 a | 5.08 ± 0.04 a | 17.57 ± 1.30 a | 5.74 ± 0.58 a | 0 ± 0 c |
| D1 | 21.6 ± 3.36 e | 3.77 ± 1.44 f | 1.82 ± 0.73 g | 1.24 ± 0.10 h | 0.52 ± 0.06 d | 3.56 ± 0.45 b | 2.80 ± 2.55 e | 1.16 ± 1.07 e | 44.98 ± 21.48 a |
| D2 | 24.2 ± 2.17 de | 4.68 ± 1.18 ef | 2.35 ± 0.76 fg | 1.60 ± 0.06 g | 0.73 ± 0.08 d | 3.78 ± 0.51 b | 4.20 ± 3.84 de | 1.34 ± 1.22 de | 37.90 ± 21.72 ab |
| D3 | 26.1 ± 1.59 cde | 6.16 ± 0.04 de | 3.10 ± 0.06 ef | 2.08 ± 0.08 f | 1.05 ± 0.08 cd | 4.94 ± 0.27 a | 7.62 ± 0.04 cd | 2.64 ± 0.15 c | 16.00 ± 0.22 bc |
| D4 | 27 ± 2.00 cd | 6.88 ± 0.12 d | 3.46 ± 0.07 de | 2.69 ± 0.16 e | 1.30 ± 0.07 cd | 4.84 ± 0.63 a | 7.90 ± 0.07 c | 2.52 ± 0.04 cd | 11.79 ± 0.27 c |
| D5 | 29.8 ± 1.30 bc | 7.70 ± 0.15 cd | 4.072 ± 0.15 cd | 3.66 ± 0.06 d | 1.80 ± 0.02 bcd | 4.98 ± 0.13 a | 10.98 ± 0.16 bc | 3.68 ± 0.13 bc | 9.35 ± 0.24 c |
| D6 | 33.8 ± 3.56 ab | 8.96 ± 0.15 bc | 4.83 ± 0.10 bc | 4.43 ± 0.06 c | 2.38 ± 0.04 bc | 4.38 ± 0.27 a | 12.14 ± 0.27 b | 4.02 ± 0.08 b | 1.36 ± 3.04 c |
| D7 | 30.6 ± 1.67 bc | 10.70 ± 0.11 b | 5.48 ± 0.12 b | 5.88 ± 0.11 b | 2.90 ± 0.02 b | 4.28 ± 0.13 a | 13.12 ± 0.15 b | 4.46 ± 0.26 b | 0.00 ± 0.00 c |
| p value | 0.05 | ||||||||
| Treatments | Severity (Days Post-Inoculation) | AUDPC | ||
|---|---|---|---|---|
| 15 | 30 | 45 | ||
| Control− | 0 h | 0 h | 0 g | 0 i |
| Control+ | 1.89 ± 0.08 a | 9.85 ± 0.21 a | 16.8 ± 0.08 a | 302.06 ± 4.18 a |
| PID1 | 1.83 ± 0.09 ab | 9.45 ± 0.13 ab | 16.62 ± 0.17 a | 293.89 ± 3.25 a |
| PID2 | 1.46 ± 0.1 b | 9.17 ± 0.17 b | 16.3 ± 0.08 ab | 279 ± 4.02 b |
| PID3 | 1.45 ± 0.13 b | 9 ± 0.18 bc | 16.01 ± 0.19 abc | 268.93 ± 2.58 c |
| PID4 | 1.23 ± 0.05 c | 8.7 ± 0.23 c | 15.65 ± 0.21 bcd | 263.63 ± 4.17 c |
| PID5 | 1.13 ± 0.05 cd | 8.63 ± 0.17 c | 15.68 ± 0.09 bcd | 253.65 ± 4.69 d |
| PID6 | 0.99 ± 0.02 d | 8.07 ± 0.13 d | 15.4 ± 0.08 cd | 253.57 ± 2.36 d |
| PID7 | 0.76 ± 0.03 e | 7.65 ± 0.1 d | 15.12 ± 0.17 d | 241.69 ± 2.4 e |
| PID8 | 0.55 ± 0.06 ef | 6.05 ± 0.19 e | 14.83 ± 0.1 de | 212.44 ± 3.02 f |
| PID9 | 0.43 ± 0.05 fg | 5.18 ± 0.3 f | 14.2 ± 0.29 e | 195.15 ± 3.46 g |
| PID10 | 0.25 ± 0.19 g | 4.25 ± 0.25 g | 12.55 ± 1.11 f | 174 ± 5.37 h |
| p value | 0.0001 | |||
| Family | Host | Local Expressions | Systemic Expressions | ELISA Detection | Report by Authors |
|---|---|---|---|---|---|
| Solanaceae | Nicotiana tabacum L. | NLL | M, D. | ANP | [4] |
| Solanaceae | Nicotiana longiflora Cav. | NLL | NS | AP | |
| Chenopodiaceae | Chenopodium giganteum D. Don. | CLL | NS | ANP | [4,15] |
| Chemopodiaceae | Chenopodium album L. | NLL, LLP | NS | ANP | [34] |
| Chenopodiaceae | Chenopodium murale L. | CLL | NS | AP | [32] |
| Solanaceae | Nicotiana glauca Graham. | CLL, CLS | NS | AP |
| Family | Host | Local Expressions | Systemic Expressions | ELISA Detection | Report by Authors |
|---|---|---|---|---|---|
| Amaranthaceae | Amaranthus hybridus L. | NS | NS | ANP | [33] |
| Convolvulaceae | Ipomoea purpurea L | NS | M, D | AP | |
| Asteraceae | Lactuca serriola L. | NS | M | AP | |
| Nyctaginaceae | Mirabilis jalapa L. | NS | M, Jl, D, AS | AP | |
| Asteraceae | Verbesina encelioides Cav. | LCS | NS | AP | |
| Labiatae | Marrubium vulgare L. | Ln, Jl | NS | AP | |
| Malvaceae | Malva neglecta Wallr. | CLS | Cn, M | AP | |
| Oxalidaceae | Oxalis latifolia Kunth | CLL | D | AP | |
| Resedaceae | Reseda luteola L. | CLL | NS | AP | |
| Araliaceae | Hedera hélix L. | CLL | NS | AP | |
| Solanaceae | Solanum eleagnifolium Cav. | NS | SC | ANP | [14] |
| Plantaginaceae | Plantago lanceolata L. | CLL | NS | AP | |
| Polygonaceae | Polygonum convolvulus L. | CLL | NS | AP | |
| Amaranthaceae | Amaranthus viridis L. | CLS | NS | AP | |
| Solanaceae | Datura quercifolia Kunth. | CLL, NLL | NS | ANP | [35] |
| Solanaceae | Datura innoxia Mill. | LLC, AS | NS | AP | |
| Asteraceae | Bidens pilosa L. | CLS | NS | AP | |
| Asteraceae | Helianthus annus L. | CLS | NS | AP | |
| Malvaceae | Malvastrum coromandelianum (L.) Garcke | CLL | NS | AP | |
| Rubiaceae | Bouvardia termifolia (Cav.) Schleter. | NS | NS | AP | |
| Solanaceae | Solanum nigrum L. | CLL | NS | ANP | [33] |
| Vitaceae | Vitis vinífera L. | CLS | NS | AP | |
| Rosaceae | Rubus idaeus L | NS | NS | AP | |
| Ranunculaceae | Clematis drummondii K. | NS | M, IY, D | AP | |
| Asteraceae | Sonchus oleraceus L. | CLL | IY, | AP | |
| Asteraceae | Titonia tubaeformis (Jacq) | CLS | NS | AP | |
| Euphorbiaceae | Ricinus comunis L. | CLL | M, D | AP | |
| Solanaceae | Solanum tuberosum L. | NS | Jl, D. | AP | [34] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vasquez Gutierrez, U.; López López, H.; Frías Treviño, G.A.; Delgado Ortiz, J.C.; Flores Olivas, A.; Aguirre Uribe, L.A.; Hernández Juarez, A. Biological Exploration and Physicochemical Characteristics of Tomato Brown Rugose Fruit Virus in Several Host Crops. Agronomy 2024, 14, 388. https://doi.org/10.3390/agronomy14020388
Vasquez Gutierrez U, López López H, Frías Treviño GA, Delgado Ortiz JC, Flores Olivas A, Aguirre Uribe LA, Hernández Juarez A. Biological Exploration and Physicochemical Characteristics of Tomato Brown Rugose Fruit Virus in Several Host Crops. Agronomy. 2024; 14(2):388. https://doi.org/10.3390/agronomy14020388
Chicago/Turabian StyleVasquez Gutierrez, Ubilfrido, Henry López López, Gustavo Alberto Frías Treviño, Juan Carlos Delgado Ortiz, Alberto Flores Olivas, Luis Alberto Aguirre Uribe, and Agustín Hernández Juarez. 2024. "Biological Exploration and Physicochemical Characteristics of Tomato Brown Rugose Fruit Virus in Several Host Crops" Agronomy 14, no. 2: 388. https://doi.org/10.3390/agronomy14020388
APA StyleVasquez Gutierrez, U., López López, H., Frías Treviño, G. A., Delgado Ortiz, J. C., Flores Olivas, A., Aguirre Uribe, L. A., & Hernández Juarez, A. (2024). Biological Exploration and Physicochemical Characteristics of Tomato Brown Rugose Fruit Virus in Several Host Crops. Agronomy, 14(2), 388. https://doi.org/10.3390/agronomy14020388