Metabolomics and DNA-Based Authentication of Two Traditional Asian Medicinal and Aromatic Species of Salvia subg. Perovskia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Sample Preparation for Qualitative Analysis
2.3. Liquid Chromatography–Mass Spectrometry Analysis
2.4. Genomic DNA Extraction
2.5. DNA Barcoding
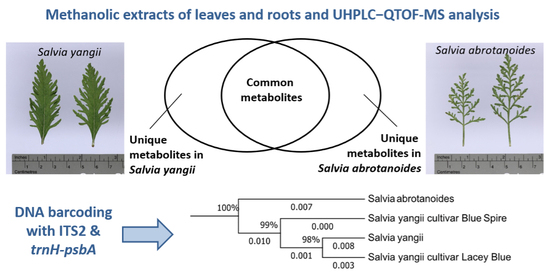
3. Results
3.1. Metabolic Analysis
3.2. Statistical Analysis of UHPLC-QTOF-MS Data
3.3. Genetic Relationship Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Flora of Pakistan. Available online: http://www.efloras.org/flora_page.aspx?flora_id=5 (accessed on 17 January 2020).
- Perveen, S.; Khan, S.B.; Malik, A.; Tareen, R.B.; Nawaz, S.A.; Choudhary, M.I. Phenolic constituents from Perovskia atriplicifolia. Nat. Prod. Res. 2006, 20, 347–353. [Google Scholar] [CrossRef]
- Perveen, S.; Malik, A.; Noor, A.T.; Tareen, R.B. Pervosides A and B, new isoferulyl glucosides from Perovskia atriplicifolia. J. Asian Nat. Prod. Res. 2008, 10, 1105–1108. [Google Scholar] [CrossRef]
- Ahmad, I.; Fozia; Waheed, A.; Tahir, N.B.; Rais, A.K. Anti-inflammatory constituents from Perovskia atriplicifolia. Pharm. Biol. 2015, 53, 1628–1631. [Google Scholar] [CrossRef]
- Rechinger, K.H. Flora Iranica; Akademische Druck-U. Verlagsanstalt: Graz, Austria, 1982. [Google Scholar]
- Mozaffarian, V. A Dictionary of Iranian Plant Names; Farhang Moaser: Tehran, Iran, 1996. [Google Scholar]
- Khaliq, S.; Volk, F.J.; Frahm, A.W. Phytochemical investigation of Perovskia abrotanoides. Planta Med. 2007, 73, 77–83. [Google Scholar] [CrossRef]
- Flora of China. Available online: http://www.efloras.org/flora_page.aspx?flora_id=2 (accessed on 17 January 2020).
- Gao, L.; Zhou, J.; Zhu, L.Y.; Zhang, J.R.; Jing, Y.X.; Zhao, J.W.; Huang, X.Z.; Li, G.P.; Jiang, Z.Y.; Xue, D.Y. Four New Diterpene Glucosides from Perovskia atriplicifolia. Chem. Biodivers. 2017, 14, 1–7. [Google Scholar] [CrossRef]
- Harley, R.M.; Atkins, S.; Budantsev, A.L.; Cantino, P.D.; Conn, B.J.; Grayer, R.; Harley, M.M.; De Kok, R.; Krestovskaja, T.; Morales, R.; et al. Labiatae. In Flowering Plants · Dicotyledons. The Families and Genera of Vascular Plants; Springer: Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- Walker, J.B.; Sytsma, K.J.; Treutlein, J.; Wink, M. Salvia (Lamiaceae) is not monophyletic: Implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. Am. J. Bot. 2004, 91, 1115–1125. [Google Scholar] [CrossRef]
- Walker, J.B.; Sytsma, K.J. Staminal evolution in the genus Salvia (Lamiaceae): Molecular phylogenetic evidence for multiple origins of the staminal lever. Ann. Bot. 2007, 100, 375–391. [Google Scholar] [CrossRef]
- Will, M.; Claßen-Bockhoff, R. Time to split Salvia s.l. (Lamiaceae)—New insights from Old World Salvia phylogeny. Mol. Phylogenet. Evol. 2017, 109, 33–58. [Google Scholar] [CrossRef]
- Drew, B.T.; González-Gallegos, J.G.; Xiang, C.L.; Kriebel, R.; Drummond, C.P.; Walker, J.B.; Sytsma, K.J. Salvia united: The greatest good for the greatest number. Taxon 2017, 66, 133–145. [Google Scholar] [CrossRef] [Green Version]
- Perveen, S.; Malik, A.; Tareen, R.B. Structural determination of atricins A and B, new triterpenes from Perovskia atriplicifolia, by 1D and 2D NMR spectroscopy. Magn. Reson. Chem. 2009, 47, 266–269. [Google Scholar] [CrossRef]
- Grubov, V.I. Plants of Central Asia: Plant Collection from China and Mongolia; Science Publishers, Inc.: Enfield, NH, USA; Plymouth, UK, 2002. [Google Scholar]
- Eisenman, S.W.; Zaurov, D.E.; Struwe, L. Medicinal Plants of Central Asia: Uzbekistan and Kyrgyzstan; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 9781461439127. [Google Scholar]
- Grant, M. RHS Plant Trials and Assessments, Perovskia; Supplementary to RHS Trials Award; Royal Horticultural Society: London, UK, 2007; pp. 1–6. [Google Scholar]
- Huxley, A. The New Royal Horticultural Society Dictionary of Gardening, 1st ed.; Nature Pub Group: London, UK, 1999; ISBN 1561592404. [Google Scholar]
- Plants of the World Online. Available online: http://www.plantsoftheworldonline.org/ (accessed on 26 January 2020).
- Tareen, R.B.; Bibi, T.; Khan, M.A.; Ahmad, M.; Zafar, M. Indigenous knowledge of folk medicine by the women of Kalat and Khuzdar regions of Balochistan, Pakistan. Pak. J. Bot. 2010, 42, 1465–1485. [Google Scholar]
- Mohammadhosseini, M.; Venditti, A.; Akbarzadeh, A. The genus Perovskia Kar.: Ethnobotany, chemotaxonomy and phytochemistry: A review. Toxin Rev. 2019, 1–22. [Google Scholar] [CrossRef]
- Baquar, S.R. Medicinal and Poisonous Plants of Pakistan, 1st ed.; Printas Press: Karachi, Pakistan, 1989. [Google Scholar]
- Jiang, Z.Y.; Yu, Y.J.; Huang, C.G.; Huang, X.Z.; Hu, Q.F.; Yang, G.Y.; Wang, H.B.; Zhang, X.Y.; Li, G.P. Icetexane diterpenoids from Perovskia atriplicifolia. Planta Med. 2015, 81, 241–246. [Google Scholar] [CrossRef] [Green Version]
- Sairafianpour, M.; Christensen, J.; Stærk, D.; Budnik, B.A.; Kharazmi, A.; Bagherzadeh, K.; Jaroszewski, J.W. Leishmanicidal, antiplasmodial, and cytotoxic activity of novel diterpenoid 1,2-quinones from Perovskia abrotanoides: New source of tanshinones. J. Nat. Prod. 2001, 64, 1398–1403. [Google Scholar] [CrossRef]
- Jaafari, M.R.; Hooshmand, S.; Samiei, A.; Hossainzadeh, H. Evaluation of leishmanicidal effect of Perovskia abrotanoides Karel. Root extract by in vitro leishmanicidal assay using promastigotes of Leishmania major. Pharmacologyonline 2007, 1, 299–303. [Google Scholar]
- Kumar, G.P.; Gupta, S.; Murugan, M.P.; Bala Singh, S. Ethnobotanical studies of Nubra valley—A cold arid zone of Himalaya. Ethnobot. Leafl. 2009, 13, 752–765. [Google Scholar]
- Moallem, S.A.; Niapour, M. Study of embryotoxicity of Perovskia abrotanoides, an adulterant in folk-medicine, during organogenesis in mice. J. Ethnopharmacol. 2008, 117, 108–114. [Google Scholar] [CrossRef]
- Ballabh, B.; Chaurasia, O.P.; Ahmed, Z.; Singh, S.B. Traditional medicinal plants of cold desert Ladakh-Used against kidney and urinary disorders. J. Ethnopharmacol. 2008, 118, 331–339. [Google Scholar] [CrossRef]
- Hosseinzadeh, H.; Amel, S. Antinociceptive Effect of the Aerial Parts of Perovskia abrotanoides Extracts in Mice. Med. J. Iran. Hosp. 2001, 4, 15–17. [Google Scholar]
- Mahboubi, M.; Kazempour, N. The antimicrobial activity of essential oil from Perovskia abrotanoides karel and its main components. Indian J. Pharm. Sci. 2009, 71, 343–347. [Google Scholar] [CrossRef] [Green Version]
- Sardashti, A.R.; Valizadeh, J.; Adhami, Y. Variation in the essential oil composition of Perovskia abrotonoides of different growth stage in Baluchestan. Middle East J. Sci. Res. 2013, 13, 781–784. [Google Scholar] [CrossRef]
- Sajjadi, S.E.; Mehregan, I.; Khatamsaz, M.; Asgari, G. Chemical composition of the essential oil of Perovskia abrotanoides Karel. growing wild in Iran. Flavour Fragr. J. 2005, 20, 445–446. [Google Scholar] [CrossRef]
- Oreizi, E.; Rahiminejad, M.R.; Asghari, G. Influence of environment on glandular trichomes and composition of essential oil of Perovskia abrotanoides Karel. Jundishapur J. Nat. Pharm. Prod. 2014, 9, e16432. [Google Scholar] [CrossRef] [Green Version]
- Pourmortazavi, S.M.; Sefidkon, F.; Hosseini, S.G. Supercritical carbon dioxide extraction of essential oils from Perovskia atriplicifolia Benth. J. Agric. Food Chem. 2003, 51, 5414–5419. [Google Scholar] [CrossRef]
- Morteza-Semnani, K. The essential oil composition of Perovskia abrotanoides from Iran. Pharm. Biol. 2004, 42, 214–216. [Google Scholar] [CrossRef]
- Jassbi, A.R.; Ahmad, V.U.; Tareen, R.B. Constituents of the essential oil of Perovskia atriplicifolia Benth. Flavour Fragr. J. 1999, 14, 38–40. [Google Scholar] [CrossRef]
- Inouye, S.; Uchida, K.; Yamaguchi, H.; Miyara, T.; Gomi, S.; Amano, M. Volatile aroma constituents of three labiatae herbs growing wild in the karakoram-himalaya district and their antifungal activity by vapor contact. J. Essent. Oil Res. 2001, 13, 68–72. [Google Scholar] [CrossRef]
- Erdemgil, F.Z.; Ilhan, S.; Korkmaz, F.; Kaplan, C.; Mercangöz, A.; Arfan, M.; Ahmad, S. Chemical composition and biological activity of the essential oil of Perovskia atriplicifolia from Pakistan. Pharm. Biol. 2007, 45, 324–331. [Google Scholar] [CrossRef]
- Dabiri, M.; Sefidkon, F. Analysis of the essential oil from aerial parts of Perovskia atriplicifolia Benth. at different stages of plant growth. Flavour Fragr. J. 2001, 16, 435–438. [Google Scholar] [CrossRef]
- Ashraf, S.N.; Zubair, M.; Rizwan, K.; Tareen, R.B.; Rasool, N.; Zia-Ul-Haq, M.; Ercisli, S. Compositional studies and biological activities of Perovskia abrotanoides kar. oils. Biol. Res. 2014, 47, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Senol, F.S.; Ślusarczyk, S.; Matkowski, A.; Pérez-Garrido, A.; Girón-Rodríguez, F.; Cerón-Carrasco, J.P.; Den-Haan, H.; Peña-García, J.; Pérez-Sánchez, H.; Domaradzki, K.; et al. Selective in vitro and in silico butyrylcholinesterase inhibitory activity of diterpenes and rosmarinic acid isolated from Perovskia atriplicifolia Benth. and Salvia glutinosa L. Phytochemistry 2017, 133, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Simmonds, M.S.J. Rosmarinic acid. Phytochemistry 2003, 62, 121–125. [Google Scholar] [CrossRef]
- Slusarczyk, S.; Topolski, J.; Domaradzki, K.; Adams, M.; Hamburger, M.; Matkowski, A. Isolation and fast selective determination of nor-abietanoid diterpenoids from Perovskia atriplicifolia roots using LC-ESI-MS/MS with multiple reaction monitoring. Nat. Prod. Commun. 2015, 10, 1149–1152. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Wu, J. Tanshinone production and isoprenoid pathways in Salvia miltiorrhiza hairy roots induced by Ag + and yeast elicitor. Plant Sci. 2005, 168, 487–491. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Inyushkina, Y.V.; Fedoreyev, S.A. Rosmarinic acid and its derivatives: Biotechnology and applications. Crit. Rev. Biotechnol. 2012, 32, 203–217. [Google Scholar] [CrossRef]
- Lu, S. Compendium of Plant Genomes; Springer Nature Switzerland AG: Cham, Switzerland, 2019; ISBN 978-3-030-24715-7. [Google Scholar]
- Wang, J.W.; Wu, J.Y. Tanshinone biosynthesis in Salvia miltiorrhiza and production in plant tissue cultures. Appl. Microbiol. Biotechnol. 2010, 88, 437–449. [Google Scholar] [CrossRef]
- Aoyagi, Y.; Takahashi, Y.; Satake, Y.; Takeya, K.; Aiyama, R.; Matsuzaki, T.; Hashimoto, S.; Kurihara, T. Cytotoxicity of abietane diterpenoids from Perovskia abrotanoides and of their semisynthetic analogues. Bioorganic Med. Chem. 2006, 14, 5285–5291. [Google Scholar] [CrossRef]
- Karimzadeh, S.M.; Moridi Farimani, M.; Amiri, M.S.; Tabefam, M.; Alilou, M.; Stuppner, H. Perovskanol, a new sesquiterpenoid with an unprecedented skeleton from Perovskia abrotanoides. Nat. Prod. Res. 2019, 1–5. [Google Scholar] [CrossRef]
- Tarawneh, A.; León, F.; Pettaway, S.; Elokely, K.M.; Klein, M.L.; Lambert, J.; Mansoor, A.; Cutler, S.J. Flavonoids from Perovskia atriplicifolia and Their in Vitro Displacement of the Respective Radioligands for Human Opioid and Cannabinoid Receptors. J. Nat. Prod. 2015, 78, 1461–1465. [Google Scholar] [CrossRef]
- Liu, W.X.; Zhao, J.W.; Zuo, A.X.; Yang, Z.; Gao, L.; Zhou, M.; Jiang, Z.Y. Two novel terpenoids from the cultured Perovskia atriplicifolia. Fitoterapia 2018, 130, 152–155. [Google Scholar] [CrossRef]
- Borges, A.; Rosa, M.; Recchia, G.; Queiroz-Silva, J.; Bressan, E.; Veasey, E. CTAB methods for DNA extraction of sweetpotato for microsatellite analysis. Sci. Agric. 2009, 66, 529–534. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, S.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Tate, J.A.; Simpson, B.B. Paraphyly of Tarasa (Malvaceae) and Diverse Origins of the Polyploid Species. Syst. Bot. 2003, 28, 723–737. [Google Scholar] [CrossRef]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.; Sánchez-García, M.; Ebersberger, I.; De Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, 465–469. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Logacheva, M.D.; Valiejo-Roman, C.M.; Pimenov, M.G. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Plant Syst. Evol. 2008, 270, 783–791. [Google Scholar] [CrossRef]
- Yan, X. Dan Shen (Salvia miltiorrhiza) in Medicine; Volume 2. Pharmacology and Quality Control; Springer: Dordrecht, The Netherlands, 2015; ISBN 978-94-017-9462-6. [Google Scholar]
- Slusarczyk, S.; Sezer Senol Deniz, F.; Abel, R.; Pecio, Ł.; Pérez-Sánchez, H.; Cerón-Carrasco, J.P.; Den-Haan, H.; Banerjee, P.; Preissner, R.; Krzyzak, E.; et al. Norditerpenoids with selective anti-cholinesterase activity from the roots of Perovskia atriplicifolia benth. Int. J. Mol. Sci. 2020, 21, 4475. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, Z.; Rahimmalek, M.; Sabzalian, M.R. Variations in Essential Oil Composition and Antioxidant Activity in Perovskia abrotanoides Kar. Collected from Different Regions in Iran. Chem. Biodivers. 2018, 15. [Google Scholar] [CrossRef] [PubMed]
- Ghaffari, Z.; Rahimmalek, M.; Sabzalian, M.R. Variation in the primary and secondary metabolites derived from the isoprenoid pathway in the Perovskia species in response to different wavelengths generated by light emitting diodes (LEDs). Ind. Crops Prod. 2019, 140, 111592. [Google Scholar] [CrossRef]
- Tabefam, M.; Farimani, M.M.; Danton, O.; Ramseyer, J.; Kaiser, M.; Ebrahimi, S.N.; Salehi, P.; Batooli, H.; Potterat, O.; Hamburger, M. Antiprotozoal Diterpenes from Perovskia abrotanoides. Planta Med. 2018, 84, 913–919. [Google Scholar] [CrossRef]
- Hu, G.X.; Takano, A.; Drew, B.T.; Liu, E.-D.; Soltis, D.E.; Soltis, P.S.; Peng, H.; Xiang, C.L. Phylogeny and staminal evolution of Salvia (Lamiaceae, Nepetoideae) in East Asia. Ann. Bot. 2018, 122, 649–668. [Google Scholar] [CrossRef]
- Pourhosseini, S.H.; Hadian, J.; Sonboli, A.; Nejad Ebrahimi, S.; Mirjalili, M.H. Genetic and Chemical Diversity in Perovskia abrotanoides Kar. (Lamiaceae) Populations Based on ISSRs Markers and Essential Oils Profile. Chem. Biodivers. 2018, 15. [Google Scholar] [CrossRef]
- Hashemifar, Z.; Rahimmalek, M. Genetic structure and variation in Perovskia abrotanoides Karel and P. atriplicifolia as revealed by molecular and morphological markers. Sci. Hortic. 2018, 230, 169–177. [Google Scholar] [CrossRef]
- Sucher, N.J.; Hennell, J.R.; Carles, M.C. Plant DNA Fingerprinting and Barcoding; Springer: Berlin/Heidelberg, Germany, 2013; ISBN 9781627033329. [Google Scholar]
- Chase, M.W.; Cowan, R.S.; Hollingsworth, P.M.; Van Den Berg, C.; Madriñán, S.; Petersen, G.; Seberg, O.; Jørgsensen, T.; Cameron, K.M.; Pedersen, N.; et al. A Proposal for a Standardised Protocol to Barcode All Land Plants. Taxon 2007, 56, 295–299. [Google Scholar] [CrossRef]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fazekas, A.J.; Burgess, K.S.; Kesanakurti, P.R.; Graham, S.W.; Newmaster, S.G.; Husband, B.C.; Percy, D.M.; Hajibabaei, M.; Barrett, S.C.H. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 2008, 3, e2802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollingsworth, P.M.; Graham, S.W.; Little, D.P. Choosing and using a plant DNA barcode. PLoS ONE 2011, 6, e19254. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The Tortoise and the hare III. Am. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef] [Green Version]
- Li, D.Z.; Gao, L.M.; Li, H.T.; Wang, H.; Ge, X.J.; Liu, J.Q.; Chen, Z.D.; Zhou, S.L.; Chen, S.L.; Yang, J.B.; et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19641–19646. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X.; et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Hollingsworth, P.M. Refining the DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19451–19452. [Google Scholar] [CrossRef] [Green Version]
- Bajpai, V.K.; Sonwal, S.; Hwang, S.-K.; Shukla, S.; Khan, I.; Dey, D.K.; Chen, L.; Simal-Gandara, J.; Xiao, J.; Huh, Y.S.; et al. Sugiol, a diterpenoid: Therapeutic actions and molecular pathways involved. Pharmacol. Res. 2020, 105313. [Google Scholar] [CrossRef]
- Masuda, T.; Inaba, Y.; Maekawa, T.; Takeda, Y.; Tamura, H.; Yamaguchi, H. Recovery mechanism of the antioxidant activity from carnosic acid quinone, an oxidized sage and rosemary antioxidant. J. Agric. Food Chem. 2002, 50, 5863–5869. [Google Scholar] [CrossRef]
- Winzer, T.; Gazda, V.; He, Z.; Kaminski, F.; Kern, M.; Larson, T.R.; Li, Y.; Meade, F.; Teodor, R.; Vaistij, F.E.; et al. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. SOM. Science 2012, 336, 1704–1708. [Google Scholar] [CrossRef] [Green Version]
- Nützmann, H.W.; Osbourn, A. Gene clustering in plant specialized metabolism. Curr. Opin. Biotechnol. 2014, 26, 91–99. [Google Scholar] [CrossRef] [Green Version]





| Primer | Sequence (5′ to 3′) | Reference |
|---|---|---|
| ITS1F | CTTGGTCATTTAGAGGAAGTAA | [54] |
| ITS4 | TCCTCCGCTTATTGATATGC | [55] |
| psbA3_f | GTTATGCATGAACGTAATGCTC | [57] |
| trnHf_05 | CGCGCATGGTGGATTCACAATCC | [56] |
| No. | Compound | RT | UV | m/z[M + H]+ | Formula | MS2 Main-Ion (Relative Intensity %) | MS2 Fragments (Relative Intensity %) | Salvia yangii | Salvia abrotanoides | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ND | 0.9 | - | 365.1123 | C13H16O10 | 169(100) | 271(45), 211(23), 151(18) | *** | *** | |
| 2 | ND | 1.7 | 237, 285, 323 | 369.1184 | C17H20O9 | 207(100) | *** | *** | ||
| 3 | Quercetin 3-rutinoside | 2.7 | 282, 342 | 611.1609 | C27H30O16 | 303(100) | 465(4) | *** | *** | HMDB0003249 |
| 4 | Luteolin-O-Hex-Dhex | 4.6 | 260, 343 | 595.1660 | C27H30O15 | 287(100) | 449(3) | *** | ** | MetFrag |
| 5 | Luteolin-O-Pen-Glucuronide | 5.1 | 260, 343 | 595.1660 | C27H30O15 | 287(100) | 395(5), 419(3) | *** | *** | MetFrag |
| 6 | Quercetin 4′-glucoside | 5.3 | 260, 348 | 465.1025 | C21H20O12 | 303(100) | *** | * | HMDB0037932 | |
| 7 | ND | 6.1 | 279, 324 | 603.2056 | C31H38O12 | 441(100) | *** | *** | ||
| 8 | Apigenin-O-Hex-Dhex | 6.7 | 282, 323 | 579.1706 | C27H31O14 | 271(100) | 433(3) | *** | ** | MetFrag |
| 9 | Hesperidin | 6.9 | 284 | 611.1970 | C27H30O16 | 303(100) | 263(5), 449(2), 369(2) | *** | * | HMDB0003265 |
| 10 | Diosmetin-O-Hex-Dhex | 7.2 | 285, 324 | 609.1815 | C28H32O15 | 301(100) | 463(3) | *** | *** | HMDB0029548 |
| 11 | Rosmarinic acid | 7.4 | 289, 328 | 361.0927 | C18H16O8 | 163(100) | 181(42), 325(15), 283(10) | *** | ** | Standard |
| 12 | ND | 8.6 | 219, 279 | 503.1915 | C26H30O10 | 279(100) | 341(32), 311(14), 291(25) | *** | ** | |
| 13 | ND | 8.8 | 219, 283, 323 | 435.1267 | C21H22O10 | 391(100) | 349(15) | ** | *** | |
| 14 | Isomargaritene | 9.2 | - | 593.1870 | C28H32O14 | 285(100) | 447(12), 429(10) | *** | * | HMDB0037415 |
| 15 | Poncirin (Isosakuranetin-7-O-beta-D-neohesperidoside) | 9.2 | - | 595.2017 | C28H34O14 | 287(100) | 269(2) | *** | * | HMDB0037487 |
| 16 | ND | 9.3 | - | 503.1920 | C26H30O10 | 279(100) | 311(90), 291(80), 251(70), 259(20) | *** | * | |
| 17 | Dihydrokaempferide 7-rhamnoside | 10.5 | 282, 320 | 449.1428 | C22H24O10 | 285(100) | ** | *** | HMDB0040561 | |
| 18 | Trihydroxy-dimethoxyflavone | 11.4 | 334 | 331.0813 | C17H14O7 | 270(100) | 298(63), 286(4) | *** | *** | HMDB0128752 |
| 19 | ND | 11.9 | - | 631.2018 | C31H34O14 | 291(100) | 323(65), 259(8) | *** | *** | |
| 20 | Isorosmanol derivative | 12.6 | - | 347.1849 | C20H26O5 | 301(100) | 273(15), 241(8), 231(7) | * | *** | MetFrag |
| 21 | Dihydroxy-methoxymethylflavone | 13.1 | 275, 333 | 315.0861 | C17H14O6 | 254(100) | 282(61), 300(13), 271(7), 285(2) | *** | * | MetFrag |
| 22 | Rosmanol | 13.4 | 288 | 347.1858 | C20H26O5 | 301(100) | 273(48), 241(27), 283(20) | * | *** | MetFrag |
| 23 | Epi-rosmanol | 14 | - | 347.1856 | C20H26O6 | 229(100) | 273(42), 283(25), 231(18), | * | *** | HMDB0035812 |
| 24 | 2-Hydroxy-3-oxo-12-oleanen-28-oic acid | 15.4 | - | 471.3471 | C30H46O4 | 453(100) | 425(42), 261(18), 201(12) | *** | HMDB0040494 | |
| 25 | 3-Oxo-12,18-ursadien-28-oic acid | 16 | - | 453.3377 | C30H44O3 | 407(100) | 201(42), 435(10), 205(10) | *** | HMDB0037065 | |
| 26 | Trihydroxy-12-ursen-28-oic acid | 16.6 | - | 489.3579 | C30H48O5 | 407(100) | 471(25), 435(14) | *** | HMDB0036961 | |
| 27 | Hydroxy-trimethoxyflavone | 16.8 | 331 | 329.1016 | C18H16O6 | 296(100) | 329(15), 284(4) | ** | *** | HMDB0040719 |
| 28 | 11-Methylrosmanol | 17 | - | 361.2020 | C21H28O4 | 329(100) | 301(48), 283(15) | * | *** | MetFrag |
| 29 | 7-Methylrosmanol | 17.5 | - | 361.2045 | C21H28O5 | 273(100) | 245(90) 301(67), 237(50), 283(41) | *** | ** | HMDB0035813 |
| 30 | Carnosol | 17.8 | 284 | 331.1903 | C20H26O4 | 285(100) | 215(45), 267(42), 243(21), 331(20) | *** | *** | HMDB0002121 |
| 31 | Carnosic acid quinone | 18.2 | 284 | 331.1949 | C20H26O5 | 285(100) | 303(42), 267(25), 243(18), 225(11), 215(10), 192(8) | *** | MetFrag | |
| 32 | 12-Hydroxy-7-oxo-8,11,13-abietatrien-18-al | 18.9 | 315.1938 | C20H26O3 | 287(100) | ** | ** | HMDB0040746 | ||
| 33 | Isorosmanol | 19.5 | - | 347.1858 | C20H26O5 | 301(100) | 231(91), 259(85), 283(45), 255(21) | *** | HMDB0036661 | |
| 34 | 11,12-Dimethylrosmanol | 19.9 | - | 375.2166 | C22H30O5 | 287(100) | 329(45), 269(22), 217(15), 191(8) | *** | *** | HMDB0040525 |
| 35 | Rosmaridiphenol | 20.1 | - | 317.2104 | C20H28O3 | 181(100) | 299(15), 273(14), 261(8) | *** | HMDB0037233 | |
| 36 | Methoxy-8,11,13-abietatrien-20,11-olide | 20.3 | - | 329.2112 | C21H28O3 | 217(100) | 191(42), 205(33), 231(17), 245(15) | *** | *** | HMDB0038391 |
| 37 | Sageone | 20.4 | - | 301.1789 | C19H24O3 | 259(100) | 241(27), 272(20), 283(18), 303(13), (216(4) | * | *** | HMDB0038684 |
| 38 | Sugiol | 21.1 | 205, 223, 282 | 301.2153 | C20H28O2 | 272(100) | 191(60), 217(51), 203(42), | *** | HMDB0036564 | |
| 39 | Trilobinol | 21.2 | 205, 223, 282 | 301.2153 | C20H28O2 | 272(100) | 191(85), 217(31), 203(22), | *** | HMDB0038702 | |
| 40 | Carnosic acid | 21.6 | - | 333.2056 | C20H28O4 | 315(100) | 287(65), 273(41), 259(12), 245(5), 233(2) | *** | * | Standard |
| 41 | Miltirone | 22.2 | - | 283.1678 | C19H22O2 | 225(100) | 268(40), 240(28), 253(20) | ** | *** | [44] |
| No. | Compound | RT | UV | m/z[M + H]+ | Formula | MS2 Main-Ion (Relative Intensity %) | MS2 Fragments (Relative Intensity %) | Salvia yangii | Salvia abrotanoides | References |
| 1 | Methoxytaxifolin | 1.3 | 283, 323 | 335.0763 | C16H14O8 | 271(100) | 253(23), 225(14), 197(8) | * | MetFrag | |
| 2 | ND | 1.8 | 217, 241, 292, 323 | 549.1594 | C26H28O13 | 531(100) | 313(8), 251(4), 295(3) | * | ||
| 3 | 2-O-p-Coumaroyltartronic acid | 2.3 | - | 267.0483 | C12H10O7 | 251(100) | 223(23), 206(15) | * | MetFrag | |
| 4 | ND | 5.4 | 287, 319 | 511.2535 | 349(100) | 331(47), 258(41), 245(25), 227(17), 211(10) | *** | *** | ||
| 5 | Salviaflaside | 5.6 | 288, 319 | 523.1439 | C24H26O13 | 163(100) | 325(45), 287(22), 361(10) | *** | *** | [65] |
| 6 | Salvianolic acid L | 6.5 | 256, 285, 315, 350 | 719.1599 | C36H30O16 | 521(100) | 295(25), 493(18), 221(12), 249(10) | *** | *** | [65] |
| 7 | Hesperidin | 6.9 | 285 | 611.1982 | C28H34O15 | 303(100) | 449(10), 263(4) | *** | HMDB03265 | |
| 8 | Chrysoeriol 7-rutinoside | 7.3 | 333 | 609.1812 | C28H32O16 | 301(100) | 463(10) | *** | HMDB37453 | |
| 9 | Rosmarinic acid | 7.5 | 288, 319 | 361.0924 | C18H16O8 | 163(100) | 181(42), 325(15), 283(10) | *** | *** | [44] |
| 10 | Melitric acid B | 7.9 | 285, 329 | 521.1062 | C27H20O11 | 295(100) | 493(22), 457(14), 329(6) | *** | HMDB0040680 | |
| 11 | 3-(3,4-dimethoxyphenyl)-5-hydroxy-7-methoxy-8-methyl-3,4-dihydro-2H-1-benzopyran-4-one | 9.8 | - | 345.1334 | C19H20O6 | 327(100) | 283(45), 268(15), 255(8), 201(4) | *** | * | HMDB0129573 |
| 12 | 3-(3,4-dimethoxyphenyl)-5-hydroxy-7-methoxy-8-methylchromen-4-one | 11 | - | 343.1170 | C19H18O6 | 325(100) | 281(51), 266(18), 251(11), 238(10), 211(4) | *** | * | HMDB0129572 |
| 13 | ND | 11.6 | 218, 339 | 315.0859 | C17H14O6 | 267(100) | 225(15) | * | ** | |
| 14 | Isograndifoliol derivative | 11.8 | - | 303.1940 | C19H26O3 | 272(100) | 203(18), 256(14), 215(7) | *** | ** | [66] |
| 15 | Tanshinone V | 12.1 | - | 315.1210 | C18H18O5 | 297(100) | 264(84), 277(21), 236(18) | *** | ** | [42] |
| 16 | ND | 12.1 | 295.1317 | 267(100) | 252(41), 237(28), 225(12) | * | *** | |||
| 17 | Dihydroxycryptotanshinon | 12.3 | - | 329.1381 | C19H20O5 | 265(100) | 283(65), 250(18), 311(14), 237(8) | *** | * | [44] |
| 18 | OH-Tanshindiol A | 12.4 | - | 331.1540 | C19H22O5 | 295(100) | 277(42), 267(41), 249(32), 235(21), 225(10) | *** | * | [65] |
| 19 | OH-cryptotanshinone | 13.5 | - | 313.1435 | C19H20O4 | 267(100) | 295(81), 280(74), 249(41), 225(25), 197(20) | *** | * | [44] |
| 20 | Oxocryptotanshinone | 13.7 | - | 311.1279 | C19H18O4 | 265(100) | 283(74), 250(51), 295(14), 237(10), 197(8) | *** | * | [44] |
| 21 | 1-βOH-cryptotanshinone | 13.8 | 220, 267 | 313.1427 | C19H20O4 | 267(100) | 295(81), 280(74), 249(41), 262(25), 197(20), 225(8) | *** | * | [44] |
| 22 | ND | 14.2 | 329.1388 | C19H20O5 | 311(100) | 285(24), 267(15), 252(8), 185(2) | *** | * | ||
| 23 | ND | 14.4 | - | 331.1898 | C19H20O4 | 311(100) | 267(14), 283(10), 225(8), 185(5) | *** | * | |
| 24 | ND | 15.5 | - | 327.1228 | C19H18O5 | 309(100) | 265(15), 250(14), 223(11), 281(8) | *** | ||
| 25 | Didehydrotanshinone IIa | 16.1 | 222, 273 | 293.1163 | C19H17O3 | 278(100) | 263(45), 247(41), 232(38), 219(18), 204(5) | *** | [44] | |
| 26 | Acetyloxy cryptotanshinone | 16.3 | - | 355.1548 | C21H22O5 | 295(100) | 280(61), 277(48), 267(38), 262(32), 249(15), 247(8) | *** | [66] | |
| 27 | Isograndifoliol | 16.7 | - | 303.1956 | C19H26O3 | 272(100) | 203(41), 215(25), 257(21), 187(18), 229(15) | *** | [66] | |
| 28 | Grandifoliol | 16.8 | - | 289.1793 | C18H24O3 | 271(100) | 256(45), 241(18) | *** | ||
| 29 | Cryptotanshinone derivative | 16.9 | - | 297.1484 | C19H20O3 | 269(100) | 254(21), 239(25), 227(18), 209(3) | *** | [42] | |
| 30 | Didehydro acetyloxy cryptotanshinone | 18.6 | - | 353.1388 | C21H20O5 | 315(100) | 293(85), 275(45), 247(28) | *** | [65] | |
| 31 | Cryptotanshinone | 18.9 | 222, 264 | 297.1469 | C19H20O3 | 269(100) | 254(41), 251(38), 279(21), 282(18) | *** | * | [44] |
| 32 | Ketoisograndifoliol | 19.4 | - | 301.1796 | C19H24O3 | 272(100) | 203(45), 215(32), 229(18), 257(12) | *** | MetFrag | |
| 33 | ND | 20.4 | - | 287.1640 | C18H23O3 | 269(100) | 254(45), 213(22), 199(18) | *** | * | |
| 34 | Tanshinone IIa | 21.7 | - | 295.1326 | C19H19O3 | 277(100) | 249(71), 262(56), 252(41), 266(20) | *** | ** | [44] |
| 35 | Miltiodiol (Arucadiol) | 22 | - | 299.1636 | C19H22O3 | 271(100) | 203(41), 253(24), 183(7) | *** | ** | [44] |
| 36 | ND | 22.1 | - | 269.1520 | C18H20O2 | 254(100) | 239(14), 225(12), 215(6) | *** | ||
| 37 | Miltirone | 22.2 | - | 283.1689 | C19H22O2 | 225(100) | 268(41), 240(32),253(28) | *** | ** | [44] |
| 38 | 6,7-Didehydroferruginol | 22.9 | - | 285.2197 | C20H28O | 271(100) | 201(12), 272(8) | *** | ** |
| Species (Voucher) | DNA Region | Accession Number (Query) | BLAST Result | Accession Number (Result) | Identity |
|---|---|---|---|---|---|
| Salvia yangii “Lacey blue” (S.OBRL.01) | ITS2 | MT599312 | Perovskia atriplicifolia (Salvia yangii) | KJ584242.1 | 99.76% |
| Salvia yangii “Lacey blue” (S.OBRL.01) | trnH-psbA | MT815871 | Salvia miltiorrhiza | KC473184.1 | 95.51% |
| Salvia yangii “Blue spire” (S.OBRL.03) | ITS2 | MT599313 | Perovskia atriplicifolia (Salvia yangii) | KJ584242.1 | 99.63% |
| Salvia yangii “Blue spire” (S.OBRL.03) | trnH-psbA | MT815872 | Salvia californica | KP852621.1 | 96.61% |
| Salvia yangii (S.OBRL.11) | ITS2 | MT599314 | Salvia yangii | DQ667223.1 | 99.62% |
| Salvia yangii (S.OBRL.11) | trnH-psbA | MT815873 | Salvia miltiorrhiza | KC473184.1 | 96.83% |
| Salvia abrotanoides (S.OBRL.12) | ITS2 | MT599315 | Salvia yangii | DQ667223.1 | 99.64 |
| Salvia abrotanoides (S.OBRL.12) | trnH-psbA | MT815874 | Salvia chionopeplica | KP852626.1 | 95.93% |
| No. | Species | Specimen Voucher | ITS2 | trnH-psbA |
|---|---|---|---|---|
| 1 | Salvia aethiopis | x142 * | DQ667272.1 | DQ667370.1 |
| 2 | Salvia amplexicaulis | C756 | MG824168.1 | MG823900.1 |
| 3 | Salvia dorystaechas | x108 * | DQ667252.1 | DQ667360.1 |
| 4 | Salvia miltiorrhiza | SMI4 | JQ934135.1 | JQ934199.1 |
| 5 | Salvia nipponica | Y. Ibaragi s. n | AB295103.1 | MG823977.1 |
| 6 | Salvia nubicola | FLPH 12-124 | MG824236.1 | MG823979.1 |
| 7 | Salvia officinalis | PS1700MT02 | FJ883522.1 | FJ513122.1 |
| 8 | Salvia paramiltiorrhiza | SPAR 3 | JQ934142.1 | JQ934206.1 |
| 9 | Salvia przewalskii | SPRZ 1 | JQ934153.1 | JQ934217.1 |
| 10 | Salvia rosmarinus | x074 * | DQ667241.1 | FJ493283.1 |
| 11 | Salvia sclarea | PS1701MT01 | FJ883529.1 | FJ513083.1 |
| 12 | Salvia koyamae | n/d | MK425922.1 | MG823960.1 |
| 13 | Salvia campanulata | PS1727MT01 | FJ883500.1 | FJ513135.1 |
| 14 | Salvia bulleyana | Yin et al. 1314 | MG824179.1 | FJ513128.1 |
| 15 | Salvia taraxacifolia | x001 * | DQ667209.1 | DQ667337.1 |
| 16 | Salvia judaica | C769 | MG824218.1 | MG823957.1 |
| 17 | Salvia lyrata | G.X. Hu QT011 | MG824225.1 | MG823966.1 |
| 18 | Salvia disjuncta | J. Walker 3018 (WIS) | MF622126.1 | MF623945.1 |
| 19 | Salvia moniliformis | Crone 15/9/00 (MJG) | MF622160.1 | MF623979.1 |
| 20 | Salvia polystachya | Mexico. Morelos F. Sazatornil 4 (MEXU) | DQ667292.1 | MF663893.1 |
| 21 | Salvia patens | x109 * | DQ667253.1 | DQ667361.1 |
| 22 | Salvia funerea | JBW 3131 | KP852812.1 | KP852638.1 |
| 23 | Salvia aristata | x170 * | DQ667280.1 | DQ667375.1 |
| 24 | Lamium maculatum | n/d | KF055055.1 | HE966679.1 |
| 25 | Lamium amplexicaule | n/d | JX073976.1 | HQ966679.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bielecka, M.; Pencakowski, B.; Stafiniak, M.; Jakubowski, K.; Rahimmalek, M.; Gharibi, S.; Matkowski, A.; Ślusarczyk, S. Metabolomics and DNA-Based Authentication of Two Traditional Asian Medicinal and Aromatic Species of Salvia subg. Perovskia. Cells 2021, 10, 112. https://doi.org/10.3390/cells10010112
Bielecka M, Pencakowski B, Stafiniak M, Jakubowski K, Rahimmalek M, Gharibi S, Matkowski A, Ślusarczyk S. Metabolomics and DNA-Based Authentication of Two Traditional Asian Medicinal and Aromatic Species of Salvia subg. Perovskia. Cells. 2021; 10(1):112. https://doi.org/10.3390/cells10010112
Chicago/Turabian StyleBielecka, Monika, Bartosz Pencakowski, Marta Stafiniak, Klemens Jakubowski, Mehdi Rahimmalek, Shima Gharibi, Adam Matkowski, and Sylwester Ślusarczyk. 2021. "Metabolomics and DNA-Based Authentication of Two Traditional Asian Medicinal and Aromatic Species of Salvia subg. Perovskia" Cells 10, no. 1: 112. https://doi.org/10.3390/cells10010112
APA StyleBielecka, M., Pencakowski, B., Stafiniak, M., Jakubowski, K., Rahimmalek, M., Gharibi, S., Matkowski, A., & Ślusarczyk, S. (2021). Metabolomics and DNA-Based Authentication of Two Traditional Asian Medicinal and Aromatic Species of Salvia subg. Perovskia. Cells, 10(1), 112. https://doi.org/10.3390/cells10010112