The Role of Chloroplast Membrane Lipid Metabolism in Plant Environmental Responses
Abstract
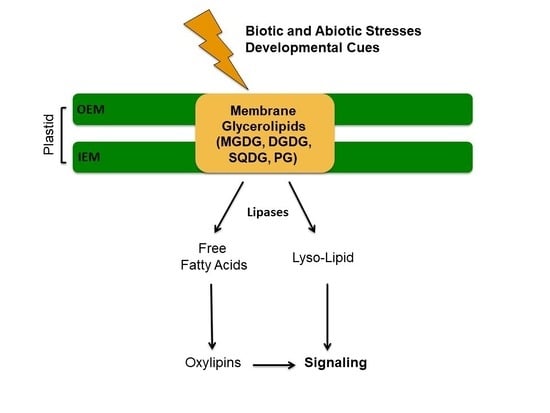
:1. Introduction
2. Chloroplast Lipid Metabolism
2.1. Biosynthesis of Lipid Precursors
2.2. Chloroplast Galactolipids
2.3. Chloroplast Anionic Lipids
2.4. The Roles of Phosphatidic Acid in Chloroplasts
2.4.1. PA Interactions with Proteins of Lipid Metabolism
2.4.2. Effects of Modifying Chloroplast PA Metabolism
2.5. Membrane Lipid Metabolism under Phosphate Limitation
2.6. Chloroplast Lipid Metabolism during Freezing
3. Connection between Thylakoid Lipid Metabolism and Oxylipin Biosynthesis
3.1. Thylakoid Lipid Specific Lipases and their Role in Biotic and Abiotic Stress
3.1.1. Temperature Variation
3.1.2. Osmotic Stress and Drought
3.1.3. Pathogen Defenses
3.1.4. Oxylipin Responses
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mackender, R.; Leech, R.M. The galactolipid, phospholipid, and fatty acid composition of the chloroplast envelope membranes of Vicia faba. L. Plant Physiol. 1974, 53, 496–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Block, M.A.; Dorne, A.-J.; Joyard, J.; Douce, R. Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. J. Biol. Chem. 1983, 258, 13281–13286. [Google Scholar] [CrossRef]
- Dörmann, P.; Benning, C. Galactolipids rule in seed plants. Trends Plant Sci. 2002, 7, 112–118. [Google Scholar] [CrossRef]
- Rocha, J.; Nitenberg, M.; Girard-Egrot, A.; Jouhet, J.; Maréchal, E.; Block, M.A.; Breton, C. Do galactolipid synthases play a key role in the biogenesis of chloroplast membranes of higher plants? Front. Plant Sci. 2018, 9, 126. [Google Scholar] [CrossRef] [Green Version]
- Holzl, G.; Dormann, P. Chloroplast lipids and their biosynthesis. Annu. Rev. Plant Biol. 2019, 70, 51–81. [Google Scholar] [CrossRef] [PubMed]
- Ohlrogge, J.B.; Kuhn, D.N.; Stumpf, P. Subcellular localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea. Proc. Natl. Acad. Sci. USA 1979, 76, 1194–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, S.W.; Zheng, J.; Zhang, Y.M.; Rock. The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 2005, 74, 791–831. [Google Scholar] [CrossRef]
- Bertrams, M.; Heinz, E. Positional specificity and fatty acid selectivity of purified sn-glycerol 3-phosphate acyltransferases from chloroplasts. Plant Physiol. 1981, 68, 653–657. [Google Scholar] [CrossRef] [Green Version]
- Kunst, L.; Somerville, C. Altered regulation of lipid biosynthesis in a mutant of Arabidopsis deficient in chloroplast glycerol-3-phosphate acyltransferase activity. Proc. Natl. Acad. Sci. USA 1988, 85, 4143–4147. [Google Scholar] [CrossRef] [Green Version]
- Yu, B.; Wakao, S.; Fan, J.; Benning, C. Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in Arabidopsis. Plant Cell Physiol. 2004, 45, 503–510. [Google Scholar] [CrossRef]
- Frentzen, M.; Heinz, E.; McKeon, T.A.; Stumpf, P.K. Specificities and selectivities of glycerol-3-phosphate acyltransferase and monoacylglycerol-3-phosphate acyltransferase from pea and spinach chloroplasts. Eur. J. Biochem. 1983, 129, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Awai, K. “Prokaryotic pathway” is not prokaryotic: Noncyanobacterial origin of the chloroplast lipid biosynthetic pathway revealed by comprehensive phylogenomic analysis. Genome Biol. Evol. 2017, 9, 3162–3178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, J.; Keegstra, K. Acyl-CoA synthetase is located in the outer membrane and acyl-CoA thioesterase in the inner membrane of pea chloroplast envelopes. Plant Physiol. 1983, 72, 735–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löhden, I.; Frentzen, M. Role of plastidial acyl-acyl carrier protein: Glycerol 3-phosphate acyltransferase and acyl-acyl carrier protein hydrolase in channelling the acyl flux through the prokaryotic and eukaryotic pathway. Planta 1988, 176, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Hares, W.; Frentzen, M. Properties of the microsomal acyl-CoA: Sn-1-acyl-glycerol-3-phosphate acyltransferase from spinach (Spinacia oleracea L.) leaves. J. Plant Physiol. 1987, 131, 49–59. [Google Scholar] [CrossRef]
- Mongrand, S.; Bessoule, J.-J.; Cabantous, F.; Cassagne, C. The C16: 3\C18: 3 fatty acid balance in photosynthetic tissues from 468 plant species. Phytochemistry 1998, 49, 1049–1064. [Google Scholar] [CrossRef]
- Block, M.A.; Dorne, A.-J.; Joyard, J.; Douce, R. The phosphatidic acid phosphatase of the chloroplast envelope is located on the inner envelope membrane. FEBS Lett. 1983, 164, 111–115. [Google Scholar] [CrossRef] [Green Version]
- Jarvis, P.; Dörmann, P.; Peto, C.A.; Lutes, J.; Benning, C.; Chory, J. Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc. Natl. Acad. Sci. USA 2000, 97, 8175–8179. [Google Scholar] [CrossRef] [Green Version]
- Miège, C.; Maréchal, E.; Shimojima, M.; Awai, K.; Block, M.A.; Ohta, H.; Takamiya, K.i.; Douce, R.; Joyard, J. Biochemical and topological properties of type A MGDG synthase, a spinach chloroplast envelope enzyme catalyzing the synthesis of both prokaryotic and eukaryotic MGDG. Eur. J. Biochem. 1999, 265, 990–1001. [Google Scholar] [CrossRef] [Green Version]
- Shimojima, M.; Ohta, H.; Iwamatsu, A.; Masuda, T.; Shioi, Y.; Takamiya, K. Cloning of the gene for monogalactosyldiacylglycerol synthase and its evolutionary origin. Proc. Natl. Acad. Sci. USA 1997, 94, 333–337. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Fan, J.; Froehlich, J.E.; Awai, K.; Benning, C. Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 2005, 17, 3094–3110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roston, R.L.; Gao, J.; Murcha, M.W.; Whelan, J.; Benning, C. TGD1,-2, and-3 proteins involved in lipid trafficking form ATP-binding cassette (ABC) transporter with multiple substrate-binding proteins. J. Biol. Chem. 2012, 287, 21406–21415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cline, K.; Keegstra, K. Galactosyltransferases involved in galactolipid biosynthesis are located in the outer membrane of pea chloroplast envelopes. Plant Physiol. 1983, 71, 366–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinz, E.; Roughan, P.G. Similarities and differences in lipid metabolism of chloroplasts isolated from 18: 3 and 16: 3 plants. Plant Physiol. 1983, 72, 273–279. [Google Scholar] [CrossRef] [Green Version]
- Andrews, J.; Ohlrogge, J.B.; Keegstra, K. Final step of phosphatidic acid synthesis in pea chloroplasts occurs in the inner envelope membrane. Plant Physiol. 1985, 78, 459–465. [Google Scholar] [CrossRef] [Green Version]
- Siebertz, M.; Heinz, E. Galactosylation of different monogalactosyldiacylglycerols by cell-free preparations from pea leaves. Hoppe-Seyler’ s Z. Physiol. Chem. 1977, 358, 27–34. [Google Scholar] [CrossRef]
- Dörmann, P.; Hoffmann-Benning, S.; Balbo, I.; Benning, C. Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 1995, 7, 1801–1810. [Google Scholar]
- Dörmann, P.; Balbo, I.; Benning, C. Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science 1999, 284, 2181–2184. [Google Scholar]
- Froehlich, J.E.; Benning, C.; Dörmann, P. The digalactosyldiacylglycerol (DGDG) synthase DGD1 is inserted into the outer envelope membrane of chloroplasts in a manner independent of the general import pathway and does not depend on direct interaction with monogalactosyldiacylglycerol synthase for DGDG biosynthesis. J. Biol. Chem. 2001, 276, 31806–31812. [Google Scholar]
- Kelly, A.A.; Froehlich, J.E.; Dörmann, P. Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 2003, 15, 2694–2706. [Google Scholar] [CrossRef] [Green Version]
- Kelly, A.A.; Kalisch, B.; Hölzl, G.; Schulze, S.; Thiele, J.; Melzer, M.; Roston, R.L.; Benning, C.; Dörmann, P. Synthesis and transfer of galactolipids in the chloroplast envelope membranes of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, 10714–10719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, J.; Mudd, J.B. Phosphatidylglycerol synthesis in pea chloroplasts. Plant Physiol. 1985, 79, 259–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haselier, A.; Akbari, H.; Weth, A.; Baumgartner, W.; Frentzen, M. Two closely related genes of Arabidopsis encode plastidial cytidinediphosphate diacylglycerol synthases essential for photoautotrophic growth. Plant Physiol. 2010, 153, 1372–1384. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Härtel, H.; Wada, H.; Hagio, M.; Yu, B.; Eakin, C.; Benning, C. The pgp1 mutant locus of Arabidopsis encodes a phosphatidylglycerolphosphate synthase with impaired activity. Plant Physiol. 2002, 129, 594–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagio, M.; Sakurai, I.; Sato, S.; Kato, T.; Tabata, S.; Wada, H. Phosphatidylglycerol is essential for the development of thylakoid membranes in Arabidopsis thaliana. Plant Cell Physiol. 2002, 43, 1456–1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babiychuk, E.; Müller, F.; Eubel, H.; Braun, H.P.; Frentzen, M.; Kushnir, S. Arabidopsis phosphatidylglycerophosphate synthase 1 is essential for chloroplast differentiation, but is dispensable for mitochondrial function. Plant J. 2003, 33, 899–909. [Google Scholar] [CrossRef]
- Lin, Y.C.; Kobayashi, K.; Hung, C.H.; Wada, H.; Nakamura, Y. Arabidopsis phosphatidylglycerophosphate phosphatase 1 involved in phosphatidylglycerol biosynthesis and photosynthetic function. Plant J. 2016, 88, 1022–1037. [Google Scholar] [CrossRef] [PubMed]
- Essigmann, B.; Güler, S.; Narang, R.A.; Linke, D.; Benning, C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1998, 95, 1950–1955. [Google Scholar] [CrossRef] [Green Version]
- Shimojima, M.; Benning, C. Native uridine 5′-diphosphate–sulfoquinovose synthase, SQD1, from spinach purifies as a 250-kDa complex. Arch. Biochem. Biophys. 2003, 413, 123–130. [Google Scholar] [CrossRef]
- Yu, B.; Xu, C.; Benning, C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA 2002, 99, 5732–5737. [Google Scholar] [CrossRef] [Green Version]
- Tietje, C.; Heinz, E. Uridine-diphospho-sulfoquinovose: Diacylglycerol sulfoquinovosyltransferase activity is concentrated in the inner membrane of chloroplast envelopes. Planta 1998, 206, 72–78. [Google Scholar] [CrossRef]
- Dorne, A.-J.; Joyard, J.; DoUCE, R. Do thylakoids really contain phosphatidylcholine? Proc. Natl. Acad. Sci. USA 1990, 87, 71–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroda, M. Phosphoinositides regulate chloroplast processes. Proc. Natl. Acad. Sci. USA 2020, 117, 9154–9156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroll, D.; Meierhoff, K.; Bechtold, N.; Kinoshita, M.; Westphal, S.; Vothknecht, U.C.; Soll, J.; Westhoff, P. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl. Acad. Sci. USA 2001, 98, 4238–4242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Kato, Y.; Otters, S.; Vothknecht, U.C.; Sakamoto, W. Essential role of VIPP1 in chloroplast envelope maintenance in Arabidopsis. Plant Cell 2012, 24, 3695–3707. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Kondo, H.; Kamikubo, H.; Kataoka, M.; Sakamoto, W. VIPP1 has a disordered c-terminal tail necessary for protecting photosynthetic membranes against stress. Plant Physiol. 2016, 171, 1983–1995. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Kusaba, M.; Tanaka, A.; Sakamoto, W. Protection of chloroplast membranes by VIPP1 rescues aberrant seedling development in arabidopsisnyc1 mutant. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Theis, J.; Gupta, T.K.; Klingler, J.; Wan, W.; Albert, S.; Keller, S.; Engel, B.D.; Schroda, M. VIPP1 rods engulf membranes containing phosphatidylinositol phosphates. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertle, A.P.; García-Cerdán, J.G.; Armbruster, U.; Shih, R.; Lee, J.J.; Wong, W.; Niyogi, K.K. A Sec14 domain protein is required for photoautotrophic growth and chloroplast vesicle formation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2020, 117, 9101–9111. [Google Scholar] [CrossRef] [Green Version]
- Dubots, E.; Botté, C.; Boudière, L.; Yamaryo-Botté, Y.; Jouhet, J.; Maréchal, E.; Block, M.A. Role of phosphatidic acid in plant galactolipid synthesis. Biochimie 2012, 94, 86–93. [Google Scholar] [CrossRef] [Green Version]
- Dubots, E.; Audry, M.; Yamaryo, Y.; Bastien, O.; Ohta, H.; Breton, C.; Marechal, E.; Block, M.A. Activation of the chloroplast monogalactosyldiacylglycerol synthase MGD1 by phosphatidic acid and phosphatidylglycerol. J. Biol. Chem. 2010, 285, 6003–6011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malherbe, A.; Block, M.A.; Joyard, J.; Douce, R. Feedback inhibition of phosphatidate phosphatase from spinach chloroplast envelope membranes by diacylglycerol. J. Biol. Chem. 1992, 267, 23546–23553. [Google Scholar] [CrossRef]
- Awai, K.; Xu, C.; Tamot, B.; Benning, C. A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proc. Natl. Acad. Sci. USA 2006, 103, 10817–10822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, B.; Benning, C. A 25-amino acid sequence of the Arabidopsis TGD2 protein is sufficient for specific binding of phosphatidic acid. J. Biol. Chem. 2009, 284, 17420–17427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Xu, C.; Benning, C. TGD4 involved in endoplasmic reticulum-to-chloroplast lipid trafficking is a phosphatidic acid binding protein. Plant J. 2012, 70, 614–623. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Fan, J.; Cornish, A.J.; Benning, C. Lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis requires the extraplastidic TGD4 protein. Plant Cell 2008, 20, 2190–2204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Anderson, N.S.; Benning, C. The phosphatidic acid binding site of the Arabidopsis trigalactosyldiacylglycerol 4 (TGD4) protein required for lipid import into chloroplasts. J. Biol. Chem. 2013, 288, 4763–4771. [Google Scholar] [CrossRef] [Green Version]
- Fritz, M.; Lokstein, H.; Hackenberg, D.; Welti, R.; Roth, M.; Zahringer, U.; Fulda, M.; Hellmeyer, W.; Ott, C.; Wolter, F.P.; et al. Channeling of eukaryotic diacylglycerol into the biosynthesis of plastidial phosphatidylglycerol. J. Biol. Chem. 2007, 282, 4613–4625. [Google Scholar] [CrossRef] [Green Version]
- Muthan, B.; Roston, R.L.; Froehlich, J.E.; Benning, C. Probing Arabidopsis chloroplast diacylglycerol pools by selectively targeting bacterial diacylglycerol kinase to suborganellar membranes. Plant Physiol. 2013, 163, 61–74. [Google Scholar] [CrossRef] [Green Version]
- Härtel, H.; Dörmann, P.; Benning, C. DGD1-independent biosynthesis of extraplastidic galactolipids after phosphate deprivation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2000, 97, 10649–10654. [Google Scholar] [CrossRef] [Green Version]
- Kelly, A.A.; Dörmann, P. DGD2, an Arabidopsis gene encoding a UDP-galactose-dependent digalactosyldiacylglycerol synthase is expressed during growth under phosphate-limiting conditions. J. Biol. Chem. 2002, 277, 1166–1173. [Google Scholar] [CrossRef] [Green Version]
- Awai, K.; Maréchal, E.; Block, M.A.; Brun, D.; Masuda, T.; Shimada, H.; Takamiya, K.-i.; Ohta, H.; Joyard, J. Two types of MGDG synthase genes, found widely in both 16: 3 and 18: 3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2001, 98, 10960–10965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, K.; Awai, K.; Nakamura, M.; Nagatani, A.; Masuda, T.; Ohta, H. Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J. 2009, 57, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Qin, C.; Welti, R.; Wang, X. Double knockouts of phospholipases Dζ1 and Dζ2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol. 2006, 140, 761–770. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Welti, R.; Wang, X. Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases Dζ1 and Dζ2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 2006, 142, 750–761. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Ramírez, A.; Oropeza-Aburto, A.; Razo-Hernández, F.; Ramírez-Chávez, E.; Herrera-Estrella, L. Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2006, 103, 6765–6770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, Y.; Koizumi, R.; Shui, G.; Shimojima, M.; Wenk, M.R.; Ito, T.; Ohta, H. Arabidopsis lipins mediate eukaryotic pathway of lipid metabolism and cope critically with phosphate starvation. Proc. Natl. Acad. Sci. USA 2009, 106, 20978–20983. [Google Scholar] [CrossRef] [Green Version]
- Eastmond, P.J.; Quettier, A.L.; Kroon, J.T.; Craddock, C.; Adams, N.; Slabas, A.R. Phosphatidic acid phosphohydrolase 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 2010, 22, 2796–2811. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, Y.; Awai, K.; Masuda, T.; Yoshioka, Y.; Takamiya, K.-i.; Ohta, H. A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 2005, 280, 7469–7476. [Google Scholar] [CrossRef] [Green Version]
- Gaude, N.; Nakamura, Y.; Scheible, W.R.; Ohta, H.; Dörmann, P. Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J. 2008, 56, 28–39. [Google Scholar] [CrossRef]
- Van Besouw, A.; Wintermans, J. Galactolipid formation in chloroplast envelopes: I. Evidence for two mechanisms in galactosylation. Biochim. Biophys. Acta-Lipids Lipid Metab. 1978, 529, 44–53. [Google Scholar] [CrossRef]
- Xu, C.; Fan, J.; Riekhof, W.; Froehlich, J.E.; Benning, C. A permease-like protein involved in ER to thylakoid lipid transfer in Arabidopsis. EMBO J. 2003, 22, 2370–2379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorne, A.-J.; Block, M.A.; Joyard, J.; Douce, R. The galactolipid: Galactolipid galactosyltransferase is located on the outer surface of the outer membrane of the chloroplast envelope. FEBS Lett. 1982, 145, 30–34. [Google Scholar] [CrossRef] [Green Version]
- Wintermans, J.; van Besouw, A.; Bögemann, G. Galactolipid formation in chloroplast envelopes II. isolation-induced changes in galactolipid composition of envelopes. Biochim. Biophys. Acta-Lipids Lipid Metab. 1981, 663, 99–107. [Google Scholar] [CrossRef]
- McKown, R.; Kuroki, G.; Warren, G. Cold responses of Arabidopsis mutants impaired in freezing tolerance. J. Exp. Bot. 1996, 47, 1919–1925. [Google Scholar] [CrossRef] [Green Version]
- Thorlby, G.; Fourrier, N.; Warren, G. The SENSITIVE TO FREEZING2 gene, required for freezing tolerance in Arabidopsis thaliana, encodes a β-glucosidase. Plant Cell 2004, 16, 2192–2203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fourrier, N.; Bédard, J.; Lopez-Juez, E.; Barbrook, A.; Bowyer, J.; Jarvis, P.; Warren, G.; Thorlby, G. A role for SENSITIVE TO FREEZING2 in protecting chloroplasts against freeze-induced damage in Arabidopsis. Plant J. 2008, 55, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Moellering, E.R.; Muthan, B.; Benning, C. Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 2010, 330, 226–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Hersh, H.L.; Benning, C. SENSITIVE TO FREEZING2 aids in resilience to salt and drought in freezing-sensitive tomato. Plant Physiol. 2016, 172, 1432–1442. [Google Scholar] [CrossRef] [Green Version]
- Lu, B.; Xu, C.; Awai, K.; Jones, A.D.; Benning, C. A small atpase protein of Arabidopsis, TGD3, involved in chloroplast lipid import. J. Biol. Chem. 2007, 282, 35945–35953. [Google Scholar] [CrossRef] [Green Version]
- Heemskerk, J.W.; Bögemann, G.; Wintermans, J. Turnover of galactolipids incorporated into chloroplast envelopes: An assay for galactolipid: Galactolipid galactosyltransferase. Biochim. Biophys. Acta-Lipids Lipid Metab. 1983, 754, 181–189. [Google Scholar] [CrossRef]
- Barnes, A.C.; Benning, C.; Roston, R.L. Chloroplast membrane remodeling during freezing stress is accompanied by cytoplasmic acidification activating SENSITIVE TO FREEZING2. Plant Physiol. 2016, 171, 2140–2149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lupette, J.; Benning, C. Human health benefits of very-long-chain polyunsaturated fatty acids from microalgae. Biochimie 2020. [Google Scholar] [CrossRef]
- Griffiths, G. Biosynthesis and analysis of plant oxylipins. Free Radic. Res. 2015, 49, 565–582. [Google Scholar] [CrossRef] [PubMed]
- Hamberg, M.; Sanz, A.; Castresana, C. α-Oxidation of fatty acids in higher plants: Identification of a pathogen-inducible oxygenase (piox) as an α-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J. Biol. Chem. 1999, 274, 24503–24513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Santino, A.; Taurino, M.; De Domenico, S.; Bonsegna, S.; Poltronieri, P.; Pastor, V.; Flors, V. Jasmonate signaling in plant development and defense response to multiple (a)biotic stresses. Plant Cell Rep. 2013, 32, 1085–1098. [Google Scholar] [CrossRef]
- Park, J.-H.; Halitschke, R.; Kim, H.B.; Baldwin, I.T.; Feldmann, K.A.; Feyereisen, R. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 2002, 31, 1–12. [Google Scholar] [CrossRef]
- Chini, A.; Monte, I.; Zamarreno, A.M.; Hamberg, M.; Lassueur, S.; Reymond, P.; Weiss, S.; Stintzi, A.; Schaller, A.; Porzel, A.; et al. An OPR3-independent pathway uses 4,5-didehydrojasmonate for jasmonate synthesis. Nat. Chem. Biol. 2018, 14, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Christensen, S.A.; Huffaker, A.; Hunter, C.T.; Alborn, H.T.; Schmelz, E.A. A maize death acid, 10-oxo-11-phytoenoic acid, is the predominant cyclopentenone signal present during multiple stress and developmental conditions. Plant Signal. Behav. 2016, 11, e1120395. [Google Scholar] [CrossRef] [Green Version]
- Grechkin, A.N. Hydroperoxide lyase and divinyl ether synthase. Prostagland. Other Lipid Mediat. 2002, 68–69, 457–470. [Google Scholar] [CrossRef]
- Grechkin, A.N.; Hamberg, M. The “heterolytic hydroperoxide lyase” is an isomerase producing a short-lived fatty acid hemiacetal. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2004, 1636, 47–58. [Google Scholar] [CrossRef]
- Fammartino, A.; Cardinale, F.; Göbel, C.; Mène-Saffrané, L.; Fournier, J.; Feussner, I.; Esquerré-Tugayé, M.-T. Characterization of a divinyl ether biosynthetic pathway specifically associated with pathogenesis in tobacco. Plant Physiol. 2007, 143, 378–388. [Google Scholar] [CrossRef] [Green Version]
- Prost, I.; Dhondt, S.; Rothe, G.; Vicente, J.; Rodriguez, M.J.; Kift, N.; Carbonne, F.; Griffiths, G.; Esquerré-Tugayé, M.-T.; Rosahl, S.; et al. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 2005, 139, 1902–1913. [Google Scholar] [CrossRef] [Green Version]
- Scala, A.; Allmann, S.; Mirabella, R.; Haring, M.A.; Schuurink, R.C. Green leaf volatiles: A plant’s multifunctional weapon against herbivores and pathogens. Int. J. Mol. Sci. 2013, 14, 17781–17811. [Google Scholar] [CrossRef] [Green Version]
- Fall, R.; Karl, T.; Hansel, A.; Jordan, A.; Lindinger, W. Volatile organic compounds emitted after leaf wounding: On-line analysis by proton-transfer-reaction mass spectrometry. J. Geophys. Res. Atmos. 1999, 104, 15963–15974. [Google Scholar] [CrossRef]
- Loughrin, J.H.; Manukian, A.; Heath, R.R.; Tumlinson, J.H. Volatiles emitted by different cotton varieties damaged by feeding beet armyworm larvae. J. Chem. Ecol. 1995, 21, 1217–1227. [Google Scholar] [CrossRef]
- Croft, K.P.C.; Juttner, F.; Slusarenko, A.J. Volatile products of the lipoxygenase pathway evolved from Phaseolus vulgaris (L.) leaves inoculated with Pseudomonas syringae pv phaseolicola. Plant Physiol. 1993, 101, 13. [Google Scholar] [CrossRef] [Green Version]
- Heiden, A.C.; Kobel, K.; Langebartels, C.; Schuh-Thomas, G.; Wildt, J. Emissions of oxygenated volatile organic compounds from plants part i: Emissions from lipoxygenase activity. J. Atmos. Chem. 2003, 45, 143–172. [Google Scholar] [CrossRef]
- Piesik, D.; Lemńczyk, G.; Skoczek, A.; Lamparski, R.; Bocianowski, J.; Kotwica, K.; Delaney, K.J. Fusarium infection in maize: Volatile induction of infected and neighboring uninfected plants has the potential to attract a pest cereal leaf beetle, Oulema melanopus. J. Plant Physiol. 2011, 168, 1534–1542. [Google Scholar] [CrossRef]
- Kishimoto, K.; Matsui, K.; Ozawa, R.; Takabayashi, J. Direct fungicidal activities of C6-aldehydes are important constituents for defense responses in Arabidopsis against Botrytis cinerea. Phytochemistry 2008, 69, 2127–2132. [Google Scholar] [CrossRef]
- Gouinguené, S.P.; Turlings, T.C.J. The effects of abiotic factors on induced volatile emissions in corn plants. Plant Physiol. 2002, 129, 1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blée, E. Impact of phyto-oxylipins in plant defense. Trends Plant Sci. 2002, 7, 315–322. [Google Scholar] [CrossRef]
- Vollenweider, S.; Weber, H.; Stolz, S.; Chételat, A.; Farmer, E.E. Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J. 2000, 24, 467–476. [Google Scholar] [CrossRef]
- Hamberg, M. An epoxy alcohol synthase pathway in higher plants: Biosynthesis of antifungal trihydroxy oxylipins in leaves of potato. Lipids 1999, 34, 1131–1142. [Google Scholar] [CrossRef]
- Blée, E. Phytooxylipins and plant defense reactions. Prog. Lipid Res. 1998, 37, 33–72. [Google Scholar] [CrossRef]
- Hamberg, M.; Hamberg, G. Peroxygenase-catalyzed fatty acid epoxidation in cereal seeds (sequential oxidation of linoleic acid into 9(s),12(s),13(s)-trihydroxy-10(e)-octadecenoic acid). Plant Physiol. 1996, 110, 807. [Google Scholar] [CrossRef] [Green Version]
- Masui, H.; Kondo, T.; Kojima, M. An antifungal compound, 9,12,13-trihydroxy-(E)-10-octadecenoic acid, from Colocasia antiquorum inoculated with Ceratocystis fimbriata. Phytochemistry 1989, 28, 2613–2615. [Google Scholar] [CrossRef]
- Parchmann, S.; Mueller, M.J. Evidence for the formation of dinor isoprostanes e1from α-linolenic acid in plants. J. Biol. Chem. 1998, 273, 32650–32655. [Google Scholar] [CrossRef] [Green Version]
- Galano, J.-M.; Lee, Y.Y.; Oger, C.; Vigor, C.; Vercauteren, J.; Durand, T.; Giera, M.; Lee, J.C.-Y. Isoprostanes, neuroprostanes and phytoprostanes: An overview of 25 years of research in chemistry and biology. Prog. Lipid Res 2017, 68, 83–108. [Google Scholar] [CrossRef]
- Stelmach, B.A.; Müller, A.; Hennig, P.; Gebhardt, S.; Schubert-Zsilavecz, M.; Weiler, E.W. A novel class of oxylipins,sn1-o-(12-oxophytodienoyl)-sn2-o-(hexadecatrienoyl)-monogalactosyl diglyceride, from Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 12832–12838. [Google Scholar] [CrossRef] [Green Version]
- Göbel, C.; Feussner, I. Methods for the analysis of oxylipins in plants. Phytochemistry 2009, 70, 1485–1503. [Google Scholar] [CrossRef]
- Mosblech, A.; Feussner, I.; Heilmann, I. Oxylipins: Structurally diverse metabolites from fatty acid oxidation. Plant Physiol. Biochem. 2009, 47, 511–517. [Google Scholar] [CrossRef]
- Chechetkin, I.R.; Mukhitova, F.K.; Blufard, A.S.; Yarin, A.Y.; Antsygina, L.L.; Grechkin, A.N. Unprecedented pathogen-inducible complex oxylipins from flax—Linolipins A and B. FEBS J. 2009, 276, 4463–4472. [Google Scholar] [CrossRef]
- Chechetkin, I.R.; Blufard, A.S.; Khairutdinov, B.I.; Mukhitova, F.K.; Gorina, S.S.; Yarin, A.Y.; Antsygina, L.L.; Grechkin, A.N. Isolation and structure elucidation of linolipins C and D, complex oxylipins from flax leaves. Phytochemistry 2013, 96, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Genva, M.; Obounou Akong, F.; Andersson, M.X.; Deleu, M.; Lins, L.; Fauconnier, M.-L. New insights into the biosynthesis of esterified oxylipins and their involvement in plant defense and developmental mechanisms. Phytochem. Rev. 2019, 18, 343–358. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-T.; Chen, L.-J.; Herrfurth, C.; Feussner, I.; Li, H.-M. Reduced biosynthesis of digalactosyldiacylglycerol, a major chloroplast membrane lipid, leads to oxylipin overproduction and phloem cap lignification in Arabidopsis. Plant Cell 2016, 28, 219–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevalier, F.; Cuyas, L.; Jouhet, J.; Gros, V.; Chiarenza, S.; Secco, D.; Whelan, J.; Seddiki, K.; Block, M.A.; Nussaume, L.; et al. Interplay between jasmonic acid, phosphate signaling and the regulation of glycerolipid homeostasis in Arabidopsis. Plant Cell Physiol. 2019, 60, 1260–1273. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.A.; Vogiatzaki, E.; Glauser, G.; Poirier, Y. Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol. 2016, 171, 632. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Du, X.; Zhou, Y.; Xie, L.; Bie, S.; Tu, L.; Zhang, N.; Yang, X.; Xiao, S.; Zhang, X. The β-ketoacyl-CoA synthase KCS13 regulates the cold response in cotton by modulating lipid and oxylipin biosynthesis. J. Exp. Bot. 2020. [Google Scholar] [CrossRef]
- Wang, K.; Durrett, T.P.; Benning, C. Functional diversity of glycerolipid acylhydrolases in plant metabolism and physiology. Prog. Lipid Res. 2019, 75, 100987. [Google Scholar] [CrossRef] [PubMed]
- Ruelland, E.; Cantrel, C.; Gawer, M.; Kader, J.-C.; Zachowski, A. Activation of phospholipases C and D is an early response to a cold exposure in Arabidopsis suspension cells. Plant Physiol. 2002, 130, 999. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Guo, Q.; Froehlich, J.E.; Hersh, H.L.; Zienkiewicz, A.; Howe, G.A.; Benning, C. Two abscisic acid-responsive plastid lipase genes involved in jasmonic acid biosynthesis in Arabidopsis thaliana. Plant Cell 2018, 30, 1006. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Moellering, E.R.; Liu, B.; Johnny, C.; Fedewa, M.; Sears, B.B.; Kuo, M.-H.; Benning, C. A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 2012, 24, 4670. [Google Scholar] [CrossRef] [Green Version]
- Du, Z.-Y.; Lucker, B.F.; Zienkiewicz, K.; Miller, T.E.; Zienkiewicz, A.; Sears, B.B.; Kramer, D.M.; Benning, C. Galactoglycerolipid lipase PGD1 is involved in thylakoid membrane remodeling in response to adverse environmental conditions in Chlamydomonas. Plant Cell 2018, 30, 447. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.-F.; Xu, L.; Tan, W.-J.; Chen, L.; Qi, H.; Xie, L.-J.; Chen, M.-X.; Liu, B.-Y.; Yu, L.-J.; Yao, N.; et al. Disruption of the Arabidopsis defense regulator genes SAG101, EDS1, and PAD4 confers enhanced freezing tolerance. Mol. Plant 2015, 8, 1536–1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Jeong, R.-D.; Venugopal, S.C.; Lapchyk, L.; Navarre, D.; Kachroo, A.; Kachroo, P. SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathog. 2011, 7, e1002318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feys, B.J.; Wiermer, M.; Bhat, R.A.; Moisan, L.J.; Medina-Escobar, N.; Neu, C.; Cabral, A.; Parker, J.E. Arabidopsis SENESCENCE-ASSOCIATED GENE101 Stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 2005, 17, 2601. [Google Scholar] [CrossRef] [Green Version]
- Higashi, Y.; Okazaki, Y.; Takano, K.; Myouga, F.; Shinozaki, K.; Knoch, E.; Fukushima, A.; Saito, K. HEAT INDUCIBLE LIPASE1 remodels chloroplastic monogalactosyldiacylglycerol by liberating α-linolenic acid in arabidopsis leaves under heat stress. Plant Cell 2018, 30, 1887. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.-Y.; Zheng, Y.; Bahn, S.C.; Pan, X.-Q.; Li, M.-Y.; Vu, H.S.; Roth, M.R.; Scheu, B.; Welti, R.; Hong, Y.-Y.; et al. The patatin-containing phospholipase A pPLAIIα modulates oxylipin formation and water loss in Arabidopsis thaliana. Mol. Plant 2012, 5, 452–460. [Google Scholar] [CrossRef] [Green Version]
- Su, H.-G.; Zhang, X.-H.; Wang, T.-T.; Wei, W.-L.; Wang, Y.-X.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Xu, Z.-S.; et al. Genome-wide identification, evolution, and expression of GDSL-type esterase/lipase gene family in soybean. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Arisz, S.A.; Munnik, T. The salt stress-induced LPA response in Chlamydomonas is produced via PLA2 hydrolysis of DGK-generated phosphatidic acid. J. Lipid Res. 2011, 52, 2012–2020. [Google Scholar] [CrossRef] [Green Version]
- Alfano, J.R.; Collmer, A. TYPE III SECRETION SYSTEM EFFECTOR PROTEINS: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopath. 2004, 42, 385–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coburn, B.; Sekirov, I.; Finlay, B.B. Type III secretion systems and disease. Clin. Microbiol. Rev. 2007, 20, 535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunnac, S.; Wilson, A.; Nuwer, J.; Kirik, A.; Baranage, G.; Mudgett, M.B. A Conserved Carboxylesterase Is a SUPPRESSOR OF AVRBST-ELICITED RESISTANCE in Arabidopsis. Plant Cell 2007, 19, 688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirik, A.; Mudgett, M.B. SOBER1 phospholipase activity suppresses phosphatidic acid accumulation and plant immunity in response to bacterial effector AvrBsT. Proc. Natl. Acad. Sci. USA 2009, 106, 20532–20537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bürger, M.; Willige, B.C.; Chory, J. A hydrophobic anchor mechanism defines a deacetylase family that suppresses host response against YopJ effectors. Nat. Commun. 2017, 8, 2201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, S.; Jayaraman, J.; Sohn, K.H. Arabidopsis thaliana SOBER1 (SUPPRESSOR OF AVRBST-ELICITED RESISTANCE 1) suppresses plant immunity triggered by multiple bacterial acetyltransferase effectors. New Phytol. 2018, 219, 324–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akoh, C.C.; Lee, G.-C.; Liaw, Y.-C.; Huang, T.-H.; Shaw, J.-F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004, 43, 534–552. [Google Scholar] [CrossRef]
- Oh, I.S.; Park, A.R.; Bae, M.S.; Kwon, S.J.; Kim, Y.S.; Lee, J.E.; Kang, N.Y.; Lee, S.; Cheong, H.; Park, O.K. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 2005, 17, 2832–2847. [Google Scholar] [CrossRef] [Green Version]
- Kwon, S.J.; Jin, H.C.; Lee, S.; Nam, M.H.; Chung, J.H.; Kwon, S.I.; Ryu, C.-M.; Park, O.K. GDSL lipase-like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis. Plant J. 2009, 58, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.G.; Kwon, S.J.; Jang, Y.J.; Chung, J.H.; Nam, M.H.; Park, O.K. GDSL lipase 1 regulates ethylene signaling and ethylene-associated systemic immunity in Arabidopsis. FEBS Lett. 2014, 588, 1652–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.G.; Kwon, S.J.; Jang, Y.J.; Nam, M.H.; Chung, J.H.; Na, Y.-C.; Guo, H.; Park, O.K. GDSL LIPASE1 modulates plant immunity through feedback regulation of ethylene signaling. Plant Physiol. 2013, 163, 1776. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.S.; Kim, B.K.; Kwon, S.J.; Jin, H.C.; Park, O.K. Arabidopsis GDSL lipase 2 plays a role in pathogen defense via negative regulation of auxin signaling. Biochem. Biophys. Res. Commun. 2009, 379, 1038–1042. [Google Scholar] [CrossRef]
- Hong, J.K.; Choi, H.W.; Hwang, I.S.; Kim, D.S.; Kim, N.H.; Choi, D.S.; Kim, Y.J.; Hwang, B.K. Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 2008, 227, 539–558. [Google Scholar] [CrossRef]
- Gao, M.; Yin, X.; Yang, W.; Lam, S.M.; Tong, X.; Liu, J.; Wang, X.; Li, Q.; Shui, G.; He, Z. GDSL lipases modulate immunity through lipid homeostasis in rice. PLoS Pathog. 2017, 13, e1006724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Froehlich, J.E.; Zienkiewicz, A.; Hersh, H.L.; Benning, C. A plastid phosphatidylglycerol lipase contributes to the export of acyl groups from plastids for seed oil biosynthesis. Plant Cell 2017, 29, 1678. [Google Scholar] [CrossRef] [PubMed]
- Aulakh, K.; Durrett, T.P. The plastid lipase PLIP1 is critical for seed viability in diacylglycerol acyltransferase1 mutant seed. Plant Physiol. 2019, 180, 1962. [Google Scholar] [CrossRef] [Green Version]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase a1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McConn, M.; Browse, J. The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 1996, 8, 403–416. [Google Scholar] [CrossRef]
- Stintzi, A.; Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 10625–10630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, P.M.; Lee, P.Y.; Biesgen, C.; Boone, J.D.; Beals, T.P.; Weiler, E.W.; Goldberg, R.B. The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 2000, 12, 1041–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, D.-X.; Feys, B.F.; James, S.; Nieto-Rostro, M.; Turner, J.G. COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280, 1091. [Google Scholar] [CrossRef] [PubMed]
- Feys, B.J.F.; Benedetti, C.E.; Penfold, C.N.; Turner, J.G. Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 1994, 6, 751. [Google Scholar] [CrossRef] [Green Version]
- Hyun, Y.; Choi, S.; Hwang, H.-J.; Yu, J.; Nam, S.-J.; Ko, J.; Park, J.-Y.; Seo, Y.S.; Kim, E.Y.; Ryu, S.B.; et al. Cooperation and functional diversification of two closely related galactolipase genes for jasmonate biosynthesis. Dev. Cell 2008, 14, 183–192. [Google Scholar] [CrossRef] [Green Version]
- Ellinger, D.; Stingl, N.; Kubigsteltig, I.I.; Bals, T.; Juenger, M.; Pollmann, S.; Berger, S.; Schuenemann, D.; Mueller, M.J. DONGLE and DEFECTIVE IN ANTHER DEHISCENCE1 lipases are not essential for wound- and pathogen-induced jasmonate biosynthesis: Redundant lipases contribute to jasmonate formation. Plant Physiol. 2010, 153, 114–127. [Google Scholar] [CrossRef] [Green Version]
- Ruduś, I.; Terai, H.; Shimizu, T.; Kojima, H.; Hattori, K.; Nishimori, Y.; Tsukagoshi, H.; Kamiya, Y.; Seo, M.; Nakamura, K.; et al. Wound-induced expression of DEFECTIVE IN ANTHER DEHISCENCE1 and DAD1-like lipase genes is mediated by both CORONATINE INSENSITIVE1-dependent and independent pathways in Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 849–860. [Google Scholar] [CrossRef]
- Bonaventure, G.; Schuck, S.; Baldwin, I.T. Revealing complexity and specificity in the activation of lipase-mediated oxylipin biosynthesis: A specific role of the Nicotiana attenuata GLA1 lipase in the activation of jasmonic acid biosynthesis in leaves and roots. Plant Cell Envrion. 2011, 34, 1507–1520. [Google Scholar] [CrossRef]
- Kallenbach, M.; Alagna, F.; Baldwin, I.T.; Bonaventure, G. Nicotiana attenuata SIPK, WIPK, NPR1, and fatty acid-amino acid conjugates participate in the induction of jasmonic acid biosynthesis by affecting early enzymatic steps in the pathway. Plant Physiol. 2010, 152, 96. [Google Scholar] [CrossRef] [Green Version]



| Lipase | Sequence ID | Organism | Substrate | |
|---|---|---|---|---|
| Cold Stress | PLIP2 | At1g02660 | A. thaliana | MGDG |
| PGD1 | Cre03.g193500 | C. reinhardtii | MGDG | |
| Freezing | SAG101 | At5g14930 | A. thaliana | TAG |
| EDS1 | At3g48090 | Not determined | ||
| PAD4 | At3g52430 | Not determined | ||
| Heat Stress | HIL1 | At4g13550 | A. thaliana | MGDG |
| Drought and Osmotic Stress | pPLAIIα | At2g26560 | A. thaliana | Several substrates |
| PLIP3 | At3g62590 | A. thaliana | PG | |
| Pathogen Defenses | SOBER1 | At4g22305 | A. thaliana | PC / PA |
| GLIP1 | At5g50990 | Synthetic esters | ||
| GLIP2 | At1g53940 | |||
| Oxylipin Responses | DAD1 | At2g44810 | A. thaliana | MGDG, PC, TAG |
| DGL | At1g06800 | A. thaliana | MGDG, DGDG, PC, TAG | |
| GLA1 | FJ821553 | N. attenuata | PC, MGDG, TAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cook, R.; Lupette, J.; Benning, C. The Role of Chloroplast Membrane Lipid Metabolism in Plant Environmental Responses. Cells 2021, 10, 706. https://doi.org/10.3390/cells10030706
Cook R, Lupette J, Benning C. The Role of Chloroplast Membrane Lipid Metabolism in Plant Environmental Responses. Cells. 2021; 10(3):706. https://doi.org/10.3390/cells10030706
Chicago/Turabian StyleCook, Ron, Josselin Lupette, and Christoph Benning. 2021. "The Role of Chloroplast Membrane Lipid Metabolism in Plant Environmental Responses" Cells 10, no. 3: 706. https://doi.org/10.3390/cells10030706
APA StyleCook, R., Lupette, J., & Benning, C. (2021). The Role of Chloroplast Membrane Lipid Metabolism in Plant Environmental Responses. Cells, 10(3), 706. https://doi.org/10.3390/cells10030706