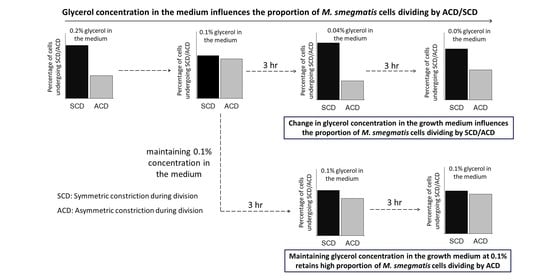
Mycobacterial Populations Partly Change the Proportions of the Cells Undergoing Asymmetric/Symmetric Divisions in Response to Glycerol Levels in Growth Medium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Growth Curve Construction
2.3. Estimation of Glycerol Concentration
2.4. Measurement of Birth-Lengths of the Cells Dividing by ACD and SCD
2.5. Calculation and Statistical Analyses of the Frequency of ACD and SCD
2.6. Effect of Glycerol Replenishment on ACD and SCD Proportions
2.7. Culturing Msm Cells in Middlebrook 7H9 Medium Containing 0.1% Glycerol and 0.05% Tween 80
2.8. Effect of Culture Supernatants of Different OD on ACD/SCD Proportions
2.9. Estimation of ACD/SCD Proportions in the Ap6A Exposed Msm MLP Cells
2.10. Total RNA Extraction from Msm MLP, 0.8 OD, and 1 h Ap6A-Treated Cultures
2.11. cDNA Preparation and Real Time PCR
2.12. Generating Msm Knockout Strains of MSMEG_2932, MSMEG_2933, and MSMEG_2936
2.13. Generating Genome Integrated Wild Type Gene Complemented Strains of the Msm Knockout Mutants MSMEG_2932 KO, MSMEG_2933 KO, and MSMEG_2936 KO
3. Results
3.1. Experimental Rationale and Strategy
3.2. Growth of Msm Cells Vis-à-Vis Glycerol Concentration in the Medium
3.3. Proportions of the Cells Dividing by ACD/SCD in Response to Glycerol Levels
3.4. Threshold Level of Glycerol for the Change of ACD/SCD Proportions
3.5. Gaussian Distribution of Daughter-to-Mother Cell-Length Ratio for ACD/SCD
3.6. Maintenance of Glycerol at 0.1% Holds the ACD:SCD Proportion at ~50:50
3.7. Starting the Culture with 0.1% Glycerol Concentration
3.8. The Influence of Glycerol in the Spent Medium on ACD/SCD Proportions
3.8.1. The Response of 0.2 OD Cells in 1.2 OD Culture Supernatant
3.8.2. The Response of 1.2 OD Cells in 0.2 OD Culture Supernatant
3.8.3. The Response of 1.2 OD Cells in 2.0 OD Culture Supernatant
3.8.4. The Response of 2.0 OD Cells in 1.2 OD Culture Supernatant
3.9. Genes Influencing the ACD/SCD Proportions in Msm Cultures
3.10. The Proportions of MSMEG_2932/2933/2936 KO Strains Dividing by ACD/SCD
4. Discussion
4.1. Link between Cell Division/Size and Nutrient Levels
4.2. Link between Cell Division and Diadenosine Polyphosphates (ApnA)
4.3. The Change in ACD/SCD Proportions and 0.1% Glycerol
4.4. Why ACD:SCD Proportion Was Studied in Glycerol and Not in Other Possible Carbon Sources?
4.5. The Relevance of ACD:SCD Proportions’ Change for Survival under Stress
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACD | asymmetric constriction during division |
| AES | allelic exchange substrate |
| CV | coefficient of variation |
| DDL | diacetyl dihydro lutidine |
| DEPC | diethyl pyrocarbonate |
| dNTP | deoxy nucleoside triphosphate |
| KO | knockout |
| MLP | mid-log phase |
| Msm | mycobacterium smegmatis |
| NCs | normal/long-sized cells |
| OD | optical density |
| PBS | phosphate buffered saline |
| PCR | polymerase chain reaction |
| PFA | paraformaldehyde |
| qPCR | quantitative polymerase chain reaction |
| RPM | revolutions per minute |
| SCD | symmetric constriction during division |
| SCs | short-sized cells |
| SDS | sodium dodecyl sulfate |
| VC | vector control |
| VRC | vanadyl ribonucleosides complex |
| XDR | extensively drug-resistant |
| XXDR | extremely drug-resistant |
References
- Ackermann, M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol. 2015, 13, 497–508. [Google Scholar] [CrossRef]
- Hallez, R.; Bellefontaine, A.F.; Letesson, J.J.; De Bolle, X. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 2004, 12, 361–365. [Google Scholar] [CrossRef]
- Imaeda, T. Ultrastructure of L-phase variants isolated from a culture of Mycobacterium phlei. J. Med. Microbiol. 1975, 8, 389–395. [Google Scholar] [CrossRef]
- Khomenko, A.G. The variability of Mycobacterium tuberculosis in patients with cavitary pulmonary tuberculosis in the course of chemotherapy. Tubercle 1987, 66, 243–253. [Google Scholar] [CrossRef]
- Smeulders, M.J.; Keer, J.; Speight, R.A.; Williams, H.D. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 1999, 181, 270–283. [Google Scholar] [CrossRef] [Green Version]
- Thanky, N.R.; Young, D.B.; Robertson, B.D. Unusual features of the cell cycle in mycobacteria: Polar-restricted growth and the snapping-model of cell division. Tuberculosis 2007, 87, 213–236. [Google Scholar] [CrossRef]
- Ryan, G.J.; Hoff, D.R.; Driver, E.R.; Voskuil, M.I.; Gonzalez-Juarrero, M.; Basaraba, R.J.; Crick, D.C.; Spencer, J.S.; Lenaerts, A.J. Multiple M. tuberculosis phenotypes in mouse and guinea pig lung tissue revealed by a dual-staining approach. PLoS ONE 2010, 5, e11108. [Google Scholar] [CrossRef] [Green Version]
- Zetola, N.M.; Modongo, C.; Moonan, P.K.; Ncube, R.; Matlhagela, K.; Sepako, E.; Collman, R.G.; Bisson, G.P. Clinical outcomes among persons with pulmonary tuberculosis caused by Mycobacterium tuberculosis isolates with phenotypic heterogeneity in results of drug-susceptibility tests. J. Infect. Dis. 2014, 209, 1754–1763. [Google Scholar] [CrossRef] [PubMed]
- Vijay, S.; Nagaraja, M.; Sebastian, J.; Ajitkumar, P. Asymmetric cell division in Mycobacterium tuberculosis and its unique features. Arch. Microbiol. 2014, 196, 157–168. [Google Scholar] [CrossRef]
- Vijay, S.; Mukkayyan, N.; Ajitkumar, P. Highly deviated asymmetric division in very low proportion of mycobacterial mid-log phase cells. Open Microbiol. J. 2014, 8, 40–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyka, W. Studies on the effect of starvation on mycobacteria. Infect. Immun. 1974, 9, 843–850. [Google Scholar] [CrossRef] [Green Version]
- Shleeva, M.O.; Mukamolova, G.V.; Young, M.; Williams, H.D.; Kaprelyants, A.S. Formation of ‘non-culturable’ cells of Mycobacterium smegmatis in stationary phase in response to growth under suboptimal conditions and their Rpf-mediated resuscitation. Microbiology 2004, 150, 1687–1697. [Google Scholar] [CrossRef] [PubMed]
- Anuchin, A.M.; Mulyukin, A.L.; Suzina, N.E.; Duda, V.I.; El-Registan, G.I.; Kaprelyants, A.S. Dormant forms of Mycobacterium smegmatis with distinct morphology. Microbiology 2009, 155, 1071–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, J.; Larsson, P.; Singh, B.; Pettersson, B.M.F.; Islam, N.M.; Sarkar, S.N.; Dasgupta, S.; Kirsebom, L.A. Sporulation in mycobacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 10781–10786. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.; Ghosh, J.; Islam, N.M.; Dasgupta, S.; Kirsebom, L.A. Growth, cell division and sporulation in mycobacteria. Antonie Leeuwenhoek 2010, 98, 165–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamont, E.A.; Bannantine, J.P.; Armien, A.; Ariyakumar, D.S.; Sreevatsan, S. Identification and characterisation of a spore-like morphotype in chronically starved Mycobacterium avium Subsp. Paratuberculosis cultures. PLoS ONE 2012, 7, e30648. [Google Scholar] [CrossRef] [Green Version]
- Slavchev, G.; Michailova, L.; Markova, N. Stress-induced L-forms of Mycobacterium bovis: A challenge to survivability. New Microbiol. 2013, 36, 157–166. [Google Scholar] [PubMed]
- Markova, N.; Slavchev, G.; Michailova, L. Unique biological properties of Mycobacterium tuberculosis L-form variants: Impact for survival under stress. Int. Microbiol. 2012, 15, 61–68. [Google Scholar] [CrossRef]
- Markova, N.; Michailova, L.; Jourdanova, M.; Kussovski, V.; Valcheva, V. Exhibition of persistent and drug tolerant L-form habit of Mycobacterium tuberculosis during infection in rats. Cent. Eur. J. Biol. 2008, 3, 407–416. [Google Scholar] [CrossRef]
- Baek, S.H.; Li, A.H.; Sessetti, C.M. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 2011, 9, e1001065. [Google Scholar] [CrossRef] [Green Version]
- Vijay, S.; Nair, R.R.; Sharan, D.; Jakkala, K.; Mukkayyan, N.; Swaminath, S.; Pradhan, A.; Joshi, N.V.; Ajitkumar, P. Mycobacterial cultures contain cell size and density specific sub-populations of cells with significant differential susceptibility to antibiotics, oxidative and nitrite stress. Front. Microbiol. 2017, 8, 463. [Google Scholar] [CrossRef] [Green Version]
- Nair, R.R.; Sharan, D.; Sebastian, J.; Swaminath, S.; Ajitkumar, P. Heterogeneity of ROS levels in antibiotic-exposed mycobacterial subpopulations confers differential susceptibility. Microbiology 2019, 165, 668–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, R.R.; Sharan, D.; Ajitkumar, P. A minor subpopulation of mycobacteria inherently produces high levels of reactive oxygen species that generate antibiotic resisters at high frequency from itself and enhance resister generation from its major kin subpopulation. Front. Microbiol. 2019, 10, 1842. [Google Scholar] [CrossRef] [Green Version]
- Dahl, J.L. Electron microscopy analysis of Mycobacterium tuberculosis cell division. FEMS Microbiol. Lett. 2004, 240, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Aldridge, B.B.; Fernandez-Suarez, M.; Heller, D.; Ambravaneswaran, V.; Irimia, D.; Toner, M.; Fortune, S.M. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 2012, 335, 100–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joyce, G.; Williams, K.J.; Robb, M.; Noens, E.; Tizzano, B.; Shahrezaei, V.; Robertson, B.D. Cell division site placement and asymmetric growth in mycobacteria. PLoS ONE 2012, 7, e44582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Middlebrook, G.; Cohn, M.L. Bacteriology of tuberculosis: Laboratory methods. Am. J. Public Health 1958, 48, 844–853. [Google Scholar] [CrossRef]
- Snapper, S.B.; Melton, R.E.; Mustafa, S.; Kieser, T.; Jacobs, W.R., Jr. Isolation and characterisation of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 1990, 4, 1911–1919. [Google Scholar] [CrossRef]
- Yanisch-Perron, C.; Vieira, J.; Messing, J. Improved M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 1985, 33, 103–119. [Google Scholar] [CrossRef]
- Csonka, L.N.; Clark, A.J. Deletions generated by the transposon Tn10 in the srl recA region of the Escherichia coli K-12 chromosome. Genetics 1979, 93, 321–343. [Google Scholar] [CrossRef]
- Widdel, F. Theory and measurement of bacterial growth. Di Dalam Grund. Mikrobiol. 2007, 4, 1–11. [Google Scholar]
- Hartman, L. Rapid determination of glycerol by the potassium periodate method. J. Appl. Chem. 1953, 3, 308–311. [Google Scholar] [CrossRef]
- Nash, T. The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem. J. 1953, 55, 416–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, D.A.; Miyada, D.S.; Nakamura, R.M. Characterisation of the periodate oxidation of glycerol and related compounds, and application toward determination of serum triglyceride concentrations. Clin. Chem. 1974, 20, 645–648. [Google Scholar] [CrossRef]
- Bondioli, P.; Bella, L.D. An alternative spectrophotometric method for the determination of free glycerol in biodiesel. Eur. J. Lipid Sci. Technol. 2005, 107, 153–157. [Google Scholar] [CrossRef]
- Kuhn, J.; Muller, H.; Salzig, D.; Czermak, P. A rapid method for an offline glycerol determination during microbial fermentation. Electron. J. Biotechnol. 2015, 18, 252–255. [Google Scholar] [CrossRef]
- Mukkayyan, N.; Sharan, D.; Ajitkumar, P. A symmetric molecule produced by mycobacteria generates cell-length asymmetry during cell-division and thereby cell-length asymmetry during cell-division and thereby cell-length heterogeneity. ACS Chem. Biol. 2018, 13, 1447–1454. [Google Scholar] [CrossRef]
- Powell, E.O. A note on Koch & Schaechter’s hypothesis about growth and fission in bacteria. J. Gen. Microbiol. 1964, 37, 231–249. [Google Scholar] [CrossRef] [Green Version]
- Marr, A.G.; Harvey, R.J.; Trentini, W.C. Growth and division of Escherichia coli. J. Bacteriol. 1966, 91, 2388–2389. [Google Scholar] [CrossRef] [Green Version]
- Treuba, F.J. On the precision and accuracy achieved by Escherichia coli cells at fission about their middle. Arch. Microbiol. 1982, 131, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Geary, R.C. The ratio of the mean deviation to the standard deviation as a test of normality. Biometrika 1935, 27, 310–332. [Google Scholar] [CrossRef]
- Wecker, E. The extraction of infectious virus nucleic acid with hot phenol. Virology 1959, 7, 241–243. [Google Scholar] [CrossRef]
- Willems, E.; Leyns, L.; Vandesompele, J. Standardisation of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008, 379, 127–129. [Google Scholar] [CrossRef]
- Wang, W.; Chen, K.; Xu, C. DNA quantification using EvaGreen and a real-time PCR instrument. Anal. Biochem. 2006, 356, 303–305. [Google Scholar] [CrossRef]
- van Kessel, J.C.; Hatfull, G.F. Recombineering in Mycobacterium tuberculosis. Nat. Meth. 2007, 4, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Bardarov, S.; Bardarov, S., Jr.; Pavelka, M.S., Jr.; Sambandamurthy, V.; Larsen, M.; Tufariello, J.; Chan, J.; Hatfull, G.; Jacobs, W.R., Jr. Specialised transduction: An efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 2002, 148, 3007–3017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibb, L.A.; Hatfull, G.F. Integration and excision of the Mycobacterium tuberculosis prophage-like element, phiRv1. Mol. Microbiol. 2002, 45, 1515–1526. [Google Scholar] [CrossRef]
- Alting-Mees, M.A.; Short, J.M. pBluescript II: Gene mapping vectors. Nucleic Acids Res. 1989, 17, 9494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ausubel, F.; Kingston, R. (Eds.) . Current Protocols in Molecular Biology; Greene Publishing: New York, NY, USA, 1987. [Google Scholar]
- Goude, R.; Parish, T. Electroporation of mycobacteria. Meth. Mol. Biol. 2009, 465, 203–215. [Google Scholar] [CrossRef]
- Mao, X.J.; Yan, M.Y.; Zhu, H.; Guo, X.P.; Sun, Y.C. Efficient and simple generation of multiple unmarked gene deletions in Mycobacterium smegmatis. Sci. Rep. 2016, 6, 22922. [Google Scholar] [CrossRef] [Green Version]
- Darch, S.E.; West, S.A.; Winzer, K.; Diggle, S.P. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc. Natl. Acad. Sci. USA 2012, 109, 8259–8263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, N.; Kim, H.; Rimbara, E.; Kato, A.; Shibayama, K.; Mori, S. Purification and functional characterisation of diadenosine 5′,5-P1,P4-tetraphosphate phosphorylases from Mycobacterium smegmatis and Mycobacterium avium. Protein Expr. Purif. 2015, 112, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Shibayama, K.; Wachino, J.I.; Arakawa, Y. Purification and molecular characterisation of a novel diadenosine 5′,5‴-P1,P4-tetraphosphate phosphorylase from Mycobacterium tuberculosis H37Rv. Protein Expr. Purif. 2010, 69, 99–105. [Google Scholar] [CrossRef]
- Zamecnik, P.C.; Stephenson, M.L.; Janeway, C.M.; Randerath, K. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 1966, 24, 91–97. [Google Scholar] [CrossRef]
- Oka, M.; Takegawa, K.; Kimura, Y. Lysyl-tRNA synthetase from Myxococcus xanthus catalyses the formation of diadenosine penta- and hexaphosphates from adenosine tetraphosphate. Arch. Biochem. Biophys. 2016, 604, 152–158. [Google Scholar]
- Dehal, P.S.; Joachimiak, M.P.; Price, M.N.; Bates, J.T.; Baumohl, J.K.; Chivian, D.; Fridland, G.D.; Huang, K.H.; Keller, K.; Novichkov, P.S.; et al. Microbesonline: An integrated portal for comparative and functional genomics. Nucleic Acids Res. 2010, 38, D396–D400. [Google Scholar] [CrossRef] [Green Version]
- Cauvin, C.; Echard, A. Phosphoinositides: Lipids with informative heads and mastermind functions in cell division. Biochim. Biophys. Acta 2014, 1851, 832–843. [Google Scholar] [CrossRef] [PubMed]
- Brenner, C.; Pace, H.C.; Garrison, P.N.; Robinson, A.K.; Rosler, A.; Liu, X.H.; Blackburn, G.M.; Croce, C.M.; Huebner, K.; Barnes, L.D. Purification and crystallisation of complexes modelling the active state of the fragile histidine triad protein. Protein Eng. 1997, 10, 1461–1463. [Google Scholar] [CrossRef] [Green Version]
- Huebner, K.; Hadaczek, P.; Siprashvili, Z.; Druck, T.; Croce, C.M. The FHIT gene, a multiple tumor suppressor gene encompassing the carcinogen sensitive chromosome fragile site, FRA3B. Biochim. Biophys. Acta 1997, 1332, 65–70. [Google Scholar] [CrossRef]
- Lima, C.D.; Klein, M.G.; Hendrickson, W.A. Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science 1997, 278, 286–290. [Google Scholar] [CrossRef]
- Le Beau, M.M.; Drabkin, H.; Glover, T.W.; Gemmill, R.; Rassool, F.V.; McKeithan, T.W.; Smith, D.I. An FHIT tumor suppressor gene? Genes Chromosomes Cancer 1998, 21, 281–289. [Google Scholar] [CrossRef]
- Kepes, F.; D’Ari, R. Involvement of FtsZ Protein in Shift-Up-Induced Division Delay in Escherichia coli. J. Bacteriol. 1987, 169, 4036–4040. [Google Scholar] [CrossRef] [Green Version]
- Chien, A.-C.; Hill, N.S.; Levin, P.A. Cell size control in bacteria. Curr. Biol. 2012, 22, R340–R349. [Google Scholar] [CrossRef] [Green Version]
- Weart, R.B.; Lee, A.H.; Chien, A.-C.; Haeusser, D.P.; Hill, N.S.; Levin, P.A. A metabolic sensor governing cell size in bacteria. Cell 2007, 130, 335–347. [Google Scholar] [CrossRef] [Green Version]
- Chien, A.-C.; Zareh, S.K.G.; Wang, Y.M.; Levin, P.A. Changes in the oligomerisation potential of the division inhibitor UgtP coordinates Bacillus subtilis cell size with nutrient availability. Mol. Microbiol. 2012, 86, 594–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, N.S.; Buske, P.J.; Shi, Y.; Levin, P.A. A Moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet. 2013, 9, e1003663. [Google Scholar] [CrossRef] [Green Version]
- Rapaport, E.; Zamecnik, P.C. Presence of diadenosine 5′,5″–P1,P4-tetraphosphate (Ap4A) in mammalian cells in levels varying widely with proliferative activity of the tissue: A possible positive “pleiotypic activator”. Proc. Natl. Acad. Sci. USA 1976, 73, 3984–3988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, A.; Moriya, S.; Ukai, H.; Nagai, K.; Wachi, M.; Yamada, Y. Diadenosine 5′,5″–P1,P4-tetraphosphate (Ap4A) controls the timing of cell division in Escherichia coli. Genes Cells 1997, 2, 401–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farr, S.B.; Arnosti, D.N.; Chamberlin, M.J.; Ames, B.N. An apaH mutation causes AppppA to accumulate and affects motility and catabolite repression in Escherichia coli. Proc. Natl. Acad. Sci. USA 1989, 86, 5010–5014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.-L.; Gengenbacher, M.; Chung, J.C.S.; Chen, S.L.; Mollenkopf, H.-J.; Kaufmann, S.H.E.; Dick, T. Developmental transcriptome of resting cell formation in Mycobacterium smegmatis. BMC Genom. 2016, 17, 837. [Google Scholar] [CrossRef] [Green Version]
- Tepper, B.S. Differences in the utilisation of glycerol and glucose by Mycobacterium phlei. J. Bacteriol. 1968, 95, 1713–1717. [Google Scholar] [CrossRef] [Green Version]
- Dingle, J.H.; Weinzirl, J. The biology of th tubercle bacillus. II. The asparagine and glycerol metabolism of the tubercle bacillus. J. Bacteriol. 1932, 23, 281–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wedum, A.G. Glycerol and carbohydrate utilisation by Mycobacterium tuberculosis. J. Bacteriol. 1936, 32, 599–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, G.J.E. The oxidation of glycerol by mycobacteria. Biochem. J. 1953, 55, 320–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priestman, M.; Thomas, P.; Robertson, B.D.; Shahrezaei, V. Mycobacteria modify their cell size control under sub-optimal carbon sources. Front. Cell Dev. Biol. 2017, 5, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mali, P.C.; Meena, L.S. Triacylglycerol: Nourishing molecule in endurance of Mycobacterium tuberculosis. J. Biosci. 2018, 43, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Farnia, P.; Masjedi, M.R.; Merza, M.A.; Tabarsi, P.; Zhavnerko, G.K.; Ibrahim, T.A.; Kuan, H.O.; Ghanavei, J.; Farnia, P.; Ranjbar, R.; et al. Growth and cell-division in extensive (XDR) and extremely drug resistant (XXDR) tuberculosis strains: Transmission and atomic force observation. Int. J. Clin. Exp. Med. 2010, 3, 308–314. [Google Scholar] [PubMed]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pradhan, A.; Mukkayyan, N.; Jakkala, K.; Ajitkumar, P. Mycobacterial Populations Partly Change the Proportions of the Cells Undergoing Asymmetric/Symmetric Divisions in Response to Glycerol Levels in Growth Medium. Cells 2021, 10, 1160. https://doi.org/10.3390/cells10051160
Pradhan A, Mukkayyan N, Jakkala K, Ajitkumar P. Mycobacterial Populations Partly Change the Proportions of the Cells Undergoing Asymmetric/Symmetric Divisions in Response to Glycerol Levels in Growth Medium. Cells. 2021; 10(5):1160. https://doi.org/10.3390/cells10051160
Chicago/Turabian StylePradhan, Atul, Nagaraja Mukkayyan, Kishor Jakkala, and Parthasarathi Ajitkumar. 2021. "Mycobacterial Populations Partly Change the Proportions of the Cells Undergoing Asymmetric/Symmetric Divisions in Response to Glycerol Levels in Growth Medium" Cells 10, no. 5: 1160. https://doi.org/10.3390/cells10051160
APA StylePradhan, A., Mukkayyan, N., Jakkala, K., & Ajitkumar, P. (2021). Mycobacterial Populations Partly Change the Proportions of the Cells Undergoing Asymmetric/Symmetric Divisions in Response to Glycerol Levels in Growth Medium. Cells, 10(5), 1160. https://doi.org/10.3390/cells10051160