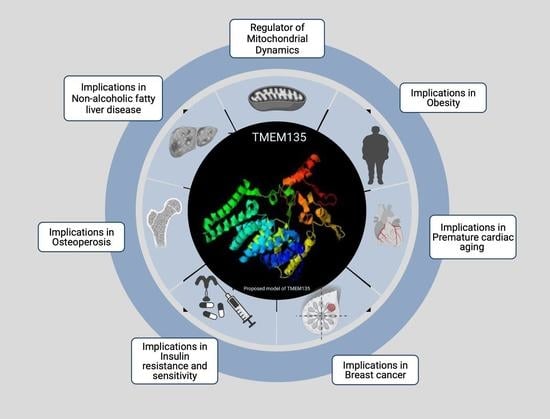
TMEM135 is a Novel Regulator of Mitochondrial Dynamics and Physiology with Implications for Human Health Conditions
Abstract
:1. The Structure and Function of Transmembrane Proteins
2. The Discovery of Transmembrane Protein 135 (TMEM135)
3. Structural Organization of the TMEM135 Gene and Protein
4. TMEM135 is a Regulator of Mitochondrial Dynamics
5. TMEM135 and Peroxisomal Transport
6. Potential Physiological Roles of TMEM135
7. Potential Role of TMEM135 as a Regulator of Calcium Dynamics
8. General Characteristics and Profiling of TMEM135 in Human Diseases
9. Perspective
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marx, S.; Maso, T.D.; Chen, J.-W.; Bury, M.; Wouters, J.; Michiels, C.; Le Calvé, B. Transmembrane (TMEM) protein family members: Poorly characterized even if essential for the metastatic process. Semin. Cancer Biol. 2020, 60, 96–106. [Google Scholar] [CrossRef]
- Fuller, C.M. Time for TMEM? J. Physiol. 2012, 590, 5931–5932. [Google Scholar] [CrossRef] [Green Version]
- Vinothkumar, K.R.; Henderson, R. Structures of membrane proteins. Q. Rev. Biophys. 2010, 43, 65–158. [Google Scholar] [CrossRef] [Green Version]
- Von Heijne, G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006, 7, 909–918. [Google Scholar] [CrossRef]
- Alberts, B. (Ed.) Molecular Biology of the Cell, 5th ed.; Alberts, B. (Ed.) Garland Science: New York, NY, USA, 2008; ISBN 978-0-8153-4105-5. [Google Scholar]
- Ruiz, M.D.L.; Kraus, R.L. Voltage-Gated Sodium Channels: Structure, Function, Pharmacology, and Clinical Indications. J. Med. Chem. 2015, 58, 7093–7118. [Google Scholar] [CrossRef]
- Abbott, G.W. KCNQs: Ligand- and Voltage-Gated Potassium Channels. Front. Physiol. 2020, 11, 583. [Google Scholar] [CrossRef] [PubMed]
- Bouza, A.A.; Isom, L.L. Voltage-Gated Sodium Channel β Subunits and Their Related Diseases. In Handbook of Experimental Pharmacology; Springer Science and Business Media LLC: New York, NY, USA, 2017; Volume 246, pp. 423–450. [Google Scholar]
- Kaplan, D.I.; Isom, L.L.; Petrou, S. Role of Sodium Channels in Epilepsy. Cold Spring Harb. Perspect. Med. 2016, 6, a022814. [Google Scholar] [CrossRef]
- Kruger, L.C.; Isom, L.L. Voltage-Gated Na+Channels: Not Just for Conduction. Cold Spring Harb. Perspect. Biol. 2016, 8, a029264. [Google Scholar] [CrossRef] [Green Version]
- Exil, V.J.; Avila, D.S.; Benedetto, A.; Exil, E.A.; Adams, M.R.; Au, C.; Aschner, M. Stressed-Induced TMEM135 Protein Is Part of a Conserved Genetic Network Involved in Fat Storage and Longevity Regulation in C. elegans. PLoS ONE 2010, 5, e14228. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.-H.; Higuchi, H.; Ikeda, S.; Macke, E.L.; Takimoto, T.; Pattnaik, B.; Liu, C.; Chu, L.-F.; Siepka, S.M.; Krentz, K.J.; et al. Mouse Tmem135 mutation reveals a mechanism involving mitochondrial dynamics that leads to age-dependent retinal pathologies. eLife 2016, 5. [Google Scholar] [CrossRef] [Green Version]
- Zaucha, J.; Heinzinger, M.; Kulandaisamy, A.; Kataka, E.; Salvádor Óscar, L.; Popov, P.; Rost, B.; Gromiha, M.M.; Zhorov, B.S.; Frishman, D. Mutations in transmembrane proteins: Diseases, evolutionary insights, prediction and comparison with globular proteins. Briefings Bioinform. 2021, 22. [Google Scholar] [CrossRef]
- Sánchez-Caballero, L.; Elurbe, D.M.; Baertling, F.; Guerrero-Castillo, S.; van der Brand, M.; van Strien, J.; van Dam, T.J.; Rodenburg, R.; Brandt, U.; Huynen, M.A.; et al. TMEM70 functions in the assembly of complexes I and V. Biochim. Biophys. Acta 2020, 1861, 148202. [Google Scholar] [CrossRef]
- Kovalčíková, J.; Vrbacký, M.; Pecina, P.; Tauchmannová, K.; Nůsková, H.; Kaplanová, V.; Brázdová, A.; Alán, L.; Eliáš, J.; Čunátová, K.; et al. TMEM70 facilitates biogenesis of mammalian ATP synthase by promoting subunit c incorporation into the rotor structure of the enzyme. FASEB J. 2019, 33, 14103–14117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahri, H.; Buratto, J.; Rojo, M.; Dompierre, J.P.; Salin, B.; Blancard, C.; Cuvellier, S.; Rose, M.; Elgaaied, A.B.A.; Tetaud, E.; et al. TMEM70 forms oligomeric scaffolds within mitochondrial cristae promoting in situ assembly of mammalian ATP synthase proton channel. Biochim. Biophys. Acta (BBA)-Bioenerg. 2021, 1868, 118942. [Google Scholar] [CrossRef]
- Vrbacky, M.; Kovalčíková, J.; Chawengsaksophak, K.; Beck, I.M.; Mráček, T.; Nůsková, H.; Sedmera, D.; Papoušek, F.; Kolar, F.; Sobol, M.; et al. Knockout of Tmem70 alters biogenesis of ATP synthase and leads to embryonal lethality in mice. Hum. Mol. Genet. 2016, 25, 4674–4685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kratochvílová, H.; Hejzlarová, K.; Vrbacky, M.; Mráček, T.; Karbanová, V.; Tesarova, M.; Gombitová, A.; Cmarko, D.; Wittig, I.; Zeman, J.; et al. Mitochondrial membrane assembly of TMEM70 protein. Mitochondrion 2014, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Magner, M.; Dvorakova, V.; Tesarova, M.; Mazurova, S.; Hansikova, H.; Zahorec, M.; Brennerova, K.; Bzduch, V.; Spiegel, R.; Horovitz, Y.; et al. TMEM70 deficiency: Long-term outcome of 48 patients. J. Inherit. Metab. Dis. 2014, 38, 417–426. [Google Scholar] [CrossRef]
- Carroll, J.; He, J.; Ding, S.; Fearnley, I.M.; Walker, J.E. TMEM70 and TMEM242 help to assemble the rotor ring of human ATP synthase and interact with assembly factors for complex I. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Mathur, A.; Sims, H.F.; Gopalakrishnan, D.; Gibson, B.; Rinaldo, P.; Vockley, J.; Hug, G.; Strauss, A.W. Molecular Heterogeneity in Very-Long-Chain Acyl-CoA Dehydrogenase Deficiency Causing Pediatric Cardiomyopathy and Sudden Death. Circulation 1999, 99, 1337–1343. [Google Scholar] [CrossRef]
- Aoyama, T.; Uchida, Y.; Kelley, R.; Marble, M.; Hofman, K.; Tonsgard, J.; Rhead, W.; Hashimoto, T. A Novel Disease with Deficiency of Mitochondrial Very-Long-Chain Acyl-CoA Dehydrogenase. Biochem. Biophys. Res. Commun. 1993, 191, 1369–1372. [Google Scholar] [CrossRef]
- Aoyama, T.; Souri, M.; Ueno, I.; Kamijo, T.; Yamaguchi, S.; Rhead, W.J.; Tanaka, K.; Hashimoto, T. Cloning of human very-long-chain acyl-coenzyme A dehydrogenase and molecular characterization of its deficiency in two patients. Am. J. Hum. Genet. 1995, 57, 273–283. [Google Scholar] [PubMed]
- Souri, M.; Aoyama, T.; Orii, K.; Yamaguchi, S.; Hashimoto, T. Mutation analysis of very-long-chain acyl-coenzyme A dehydrogenase (VLCAD) deficiency: Identification and characterization of mutant VLCAD cDNAs from four patients. Am. J. Hum. Genet. 1996, 58, 97–106. [Google Scholar] [PubMed]
- Bertrand, C.; Largillière, C.; Zabot, M.T.; Mathieu, M.; Vianey-Saban, C. Very long chain acyl-CoA dehydrogenase deficiency: Identification of a new inborn error of mitochondrial fatty acid oxidation in fibroblasts. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 1993, 1180, 327–329. [Google Scholar] [CrossRef]
- Exil, V.J.; Roberts, R.L.; Sims, H.; McLaughlin, J.E.; Malkin, R.A.; Gardner, C.D.; Ni, G.; Rottman, J.N.; Strauss, A.W. Very-Long-Chain Acyl-Coenzyme A Dehydrogenase Deficiency in Mice. Circ. Res. 2003, 93, 448–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheideler, M.; Elabd, C.; Zaragosi, L.-E.; Chiellini, C.; Hackl, H.; Sanchez-Cabo, F.; Yadav, S.; Duszka, K.; Friedl, G.; Papak, C.; et al. Comparative transcriptomics of human multipotent stem cells during adipogenesis and osteoblastogenesis. BMC Genom. 2008, 9, 340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antebi, A.; Culotti, J.; Hedgecock, E. daf-12 regulates developmental age and the dauer alternative in C. elegans. Development 1998, 125, 1191–1205. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, S.; Ruvkun, G. daf-2, daf-16 and daf-23: Genetically interacting genes controlling Dauer formation in C. elegans. Genetics 1994, 137, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Dorman, J.B.; Rodan, A.; Kenyon, C. daf-16: An HNF-3/forkhead Family Member That Can Function to Double the Life-Span of C. elegans. Science 1997, 278, 1319–1322. [Google Scholar] [CrossRef] [Green Version]
- Ogg, S.; Paradis, S.; Gottlieb, S.; Patterson, G.I.; Lee, L.; Tissenbaum, H.A.; Ruvkun, G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nat. Cell Biol. 1997, 389, 994–999. [Google Scholar] [CrossRef]
- Paradis, S.; Ruvkun, G. C. elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998, 12, 2488–2498. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.S.; Kennedy, S.; Tolonen, A.; Ruvkun, G. DAF-16 Target Genes That Control C. elegans Life-Span and Metabolism. Science 2003, 300, 644–647. [Google Scholar] [CrossRef] [Green Version]
- McElwee, J.; Bubb, K.; Thomas, J.H. Transcriptional outputs of the C. elegans forkhead protein DAF-16. Aging Cell 2003, 2, 111–121. [Google Scholar] [CrossRef] [Green Version]
- Kimura, K.; Tissenbaum, H.A.; Liu, Y.; Ruvkun, G. daf-2, an Insulin Receptor-Like Gene That Regulates Longevity and Diapause in C. elegans. Science 1997, 277, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosca, M.; Hoppel, C.L. Mitochondrial dysfunction in heart failure. Hear. Fail. Rev. 2013, 18, 607–622. [Google Scholar] [CrossRef] [Green Version]
- Liesa, M.; Palacín, M.; Zorzano, A. Mitochondrial Dynamics in Mammalian Health and Disease. Physiol. Rev. 2009, 89, 799–845. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Chan, D.C. Physiological functions of mitochondrial fusion. Ann. N. Y. Acad. Sci. 2010, 1201, 21–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naon, D.; Zaninello, M.; Giacomello, M.; Varanita, T.; Grespi, F.; Lakshminaranayan, S.; Serafini, A.; Semenzato, M.; Herkenne, S.; Hernández-Alvarez, M.I.; et al. Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum–mitochondria tether. Proc. Natl. Acad. Sci. USA 2016, 113, 11249–11254. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, J.P.; Ivanova, S.; Wandelmer, J.S.; Martínez-Cristóbal, P.; Noguera, M.; Sancho, A.; Díaz-Ramos, A.; Hernández-Alvarez, M.I.; Sebastián, D.; Mauvezin, C.; et al. Mfn2 modulates the UPR and mitochondrial function via repression of PERK. EMBO J. 2013, 32, 2348–2361. [Google Scholar] [CrossRef] [Green Version]
- Sebastián, D.; Sorianello, E.; Segalés, J.; Irazoki, A.; Ruiz-Bonilla, V.; Sala, D.; Planet, E.; Berenguer-Llergo, A.; Muñoz, J.P.; Sánchez-Feutrie, M.; et al. Mfn2 deficiency links age-related sarcopenia and impaired autophagy to activation of an adaptive mitophagy pathway. EMBO J. 2016, 35, 1677–1693. [Google Scholar] [CrossRef]
- Favaro, G.; Romanello, V.; Varanita, T.; Desbats, M.A.; Morbidoni, V.; Tezze, C.; Albiero, M.; Canato, M.; Gherardi, G.; De Stefani, D.; et al. DRP1-mediated mitochondrial shape controls calcium homeostasis and muscle mass. Nat. Commun. 2019, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dulac, M.; Leduc-Gaudet, J.; Reynaud, O.; Ayoub, M.; Guérin, A.; Finkelchtein, M.; Na Hussain, S.; Gouspillou, G. Drp1 knockdown induces severe muscle atrophy and remodelling, mitochondrial dysfunction, autophagy impairment and denervation. J. Physiol. 2020, 598, 3691–3710. [Google Scholar] [CrossRef] [PubMed]
- Scott, I.; Youle, R.J. Mitochondrial fission and fusion. Essays Biochem. 2010, 47, 85–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.-H.; Bhute, V.J.; Higuchi, H.; Ikeda, S.; Palecek, S.P.; Ikeda, A. Metabolic alterations caused by the mutation and overexpression of the Tmem135 gene. Exp. Biol. Med. 2020, 245, 1571–1583. [Google Scholar] [CrossRef]
- Pereira, R.O.; Tadinada, S.M.; Zasadny, F.M.; Oliveira, K.J.; Pires, K.M.P.; Olvera, A.; Jeffers, J.; Souvenir, R.; McGlauflin, R.; Seei, A.; et al. OPA 1 deficiency promotes secretion of FGF 21 from muscle that prevents obesity and insulin resistance. EMBO J. 2017, 36, 2126–2145. [Google Scholar] [CrossRef]
- Ren, L.; Chen, X.; Chen, X.; Li, J.; Cheng, B.; Xia, J. Mitochondrial Dynamics: Fission and Fusion in Fate Determination of Mesenchymal Stem Cells. Front. Cell Dev. Biol. 2020, 8, 580070. [Google Scholar] [CrossRef]
- Cipolat, S.; De Brito, O.M.; Zilio, B.D.; Scorrano, L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. USA 2004, 101, 15927–15932. [Google Scholar] [CrossRef] [Green Version]
- Varanita, T.; Soriano, M.E.; Romanello, V.; Zaglia, T.; Quintana-Cabrera, R.; Semenzato, M.; Menabò, R.; Costa, V.; Civiletto, G.; Pesce, P.; et al. The Opa1-Dependent Mitochondrial Cristae Remodeling Pathway Controls Atrophic, Apoptotic, and Ischemic Tissue Damage. Cell Metab. 2015, 21, 834–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietrich, M.O.; Liu, Z.-W.; Horvath, T.L. Mitochondrial Dynamics Controlled by Mitofusins Regulate Agrp Neuronal Activity and Diet-Induced Obesity. Cell 2013, 155, 188–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewis, S.; Takimoto, T.; Mehrvar, S.; Higuchi, H.; Doebley, A.-L.; Stokes, G.; Sheibani, N.; Ikeda, S.; Ranji, M.; Ikeda, A. The effect of Tmem135 overexpression on the mouse heart. PLoS ONE 2018, 13, e0201986. [Google Scholar] [CrossRef] [Green Version]
- Faust, J.; Verma, A.; Peng, C.; McNew, J.A. An Inventory of Peroxisomal Proteins and Pathways in Drosophila melanogaster. Traffic 2012, 13, 1378–1392. [Google Scholar] [CrossRef] [Green Version]
- Cipolla, C.M.; Lodhi, I.J. Peroxisomal Dysfunction in Age-Related Diseases. Trends Endocrinol. Metab. 2017, 28, 297–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renquist, B.J.; Madanayake, T.W.; Hennebold, J.D.; Ghimire, S.; Geisler, C.E.; Xu, Y.; Bogan, R.L. TMEM135 Is an LXR-Inducible Regulator of Peroxisomal Metabolism. Cell Biology 2018, 334979. [Google Scholar] [CrossRef]
- Teraoka, S.N.; Bernstein, J.L.; Reiner, A.S.; Haile, R.W.; Bernstein, L.; Lynch, C.F.; Malone, K.E.; Stovall, M.; Capanu, M.; Liang, X.; et al. Single nucleotide polymorphisms associated with risk for contralateral breast cancer in the Women’s Environment, Cancer, and Radiation Epidemiology (WECARE) Study. Breast Cancer Res. 2011, 13, R114. [Google Scholar] [CrossRef] [Green Version]
- Elbein, S.C.; Kern, P.A.; Rasouli, N.; Yao-Borengasser, A.; Sharma, N.K.; Das, S.K. Global Gene Expression Profiles of Subcutaneous Adipose and Muscle From Glucose-Tolerant, Insulin-Sensitive, and Insulin-Resistant Individuals Matched for BMI. Diabetes 2011, 60, 1019–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.; Li, W.-D.; Glessner, J.; Grant, S.; Hakonarson, H.; Price, R.A. Large Copy-Number Variations Are Enriched in Cases With Moderate to Extreme Obesity. Diabetes 2010, 59, 2690–2694. [Google Scholar] [CrossRef] [Green Version]
- Rieusset, J. The role of endoplasmic reticulum-mitochondria contact sites in the control of glucose homeostasis: An update. Cell Death Dis. 2018, 9, 388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tubbs, E.; Chanon, S.; Robert, M.; Bendridi, N.; Bidaux, G.; Chauvin, M.-A.; Ji-Cao, J.; Durand, C.; Gauvrit-Ramette, D.; Vidal, H.; et al. Disruption of Mitochondria-Associated Endoplasmic Reticulum Membrane (MAM) Integrity Contributes to Muscle Insulin Resistance in Mice and Humans. Diabetes 2018, 67, 636–650. [Google Scholar] [CrossRef] [Green Version]
- Tubbs, E.; Theurey, P.; Vial, G.; Bendridi, N.; Bravard, A.; Chauvin, M.-A.; Ji-Cao, J.; Zoulim, F.; Bartosch, B.; Ovize, M.; et al. Mitochondria-Associated Endoplasmic Reticulum Membrane (MAM) Integrity Is Required for Insulin Signaling and Is Implicated in Hepatic Insulin Resistance. Diabetes 2014, 63, 3279–3294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzmicic, J.; del Campo, A.; López-Crisosto, C.; Morales, P.E.; Pennanen, C.; Bravo-Sagua, R.; Hechenleitner, J.; Zepeda, R.; Castro, P.F.; Verdejo, H.E.; et al. Dinámica mitocondrial: Un potencial nuevo blanco terapéutico para la insuficiencia cardiaca. Rev. Española Cardiol. 2011, 64, 916–923. [Google Scholar] [CrossRef] [PubMed]
- Correa-Rodríguez, M.; Viatte, S.; Massey, J.; Schmidt-RioValle, J.; Rueda-Medina, B.; Orozco, G. Analysis of SNP-SNP interactions and bone quantitative ultrasound parameter in early adulthood. BMC Med Genet. 2017, 18, 107. [Google Scholar] [CrossRef] [Green Version]
- Mullin, B.H.; Zhao, J.H.; Brown, S.J.; Perry, J.R.; Luan, J.; Zheng, H.-F.; Langenberg, C.; Dudbridge, F.; Scott, R.; Wareham, N.J.; et al. Genome-wide association study meta-analysis for quantitative ultrasound parameters of bone identifies five novel loci for broadband ultrasound attenuation. Hum. Mol. Genet. 2017, 26, 2791–2802. [Google Scholar] [CrossRef] [Green Version]
- Breckenridge, D.G.; Stojanovic, M.; Marcellus, R.C.; Shore, G.C. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J. Cell Biol. 2003, 160, 1115–1127. [Google Scholar] [CrossRef]
- Chu, B.-B.; Liao, Y.-C.; Qi, W.; Xie, C.; Du, X.; Wang, J.; Yang, H.; Miao, H.-H.; Li, B.-L.; Song, B.-L. Cholesterol Transport through Lysosome-Peroxisome Membrane Contacts. Cell 2015, 161, 291–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Silva, B.S.; DiGiovanni, L.; Kumar, R.; Carmichael, R.E.; Kim, P.K.; Schrader, M. Maintaining social contacts: The physiological relevance of organelle interactions. Biochim. Biophys. Acta (BBA) Bioenerg. 2020, 1867, 118800. [Google Scholar] [CrossRef] [PubMed]
- Maharjan, Y.; Lee, J.N.; Kwak, S.A.; Dutta, R.K.; Park, C.; Choe, S.; Park, R. TMEM135 regulates primary ciliogenesis through modulation of intracellular cholesterol distribution. EMBO Rep. 2020, 21, e48901. [Google Scholar] [CrossRef] [PubMed]
- Schmit, K.; Michiels, C. TMEM Proteins in Cancer: A Review. Front. Pharmacol. 2018, 9, 1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [Green Version]
- Hittelman, K.J.; Fairhurst, A.S.; Smith, R.E. Calcium accumulation as a parameter of energy metabolism in mitochondria of brown adipose tissue. Proc. Natl. Acad. Sci. USA 1967, 58, 697–702. [Google Scholar] [CrossRef] [Green Version]
- Golic, I.; Veličković, K.; Markelic, M.; Stancic, A.; Jankovic, A.; Vucetic, M.; Otasevic, V.; Buzadzic, B.; Korac, B.; Korać, A. Calcium-induced alteration of mitochondrial morphology and mitochondrial-endoplasmic reticulum contacts in rat brown adipocytes. Eur. J. Histochem. 2014, 58, 2377. [Google Scholar] [CrossRef] [Green Version]
- Otera, H.; Ishihara, N.; Mihara, K. New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta (BBA)-Bioenerg. 2013, 1833, 1256–1268. [Google Scholar] [CrossRef] [Green Version]
- Giacomello, M.; Pellegrini, L. The coming of age of the mitochondria–ER contact: A matter of thickness. Cell Death Differ. 2016, 23, 1417–1427. [Google Scholar] [CrossRef]
- Vance, J.E. MAM (mitochondria-associated membranes) in mammalian cells: Lipids and beyond. Biochim. Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2014, 1841, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Rowland, A.A.; Voeltz, G.K. Endoplasmic reticulum–mitochondria contacts: Function of the junction. Nat. Rev. Mol. Cell Biol. 2012, 13, 607–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewenter, M.; Von Der Lieth, A.; Katus, H.A.; Backs, J. Calcium Signaling and Transcriptional Regulation in Cardiomyocytes. Circ. Res. 2017, 121, 1000–1020. [Google Scholar] [CrossRef] [PubMed]
- West, A.E.; Chen, W.G.; Dalva, M.B.; Dolmetsch, R.E.; Kornhauser, J.M.; Shaywitz, A.J.; Takasu, M.A.; Tao, X.; Greenberg, M.E. Calcium regulation of neuronal gene expression. Proc. Natl. Acad. Sci. USA 2001, 98, 11024–11031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shambharkar, P.B.; Bittinger, M.; Latario, B.; Xiong, Z.; Bandyopadhyay, S.; Davis, V.; Lin, V.; Yang, Y.; Valdez, R.; Labow, M.A. TMEM203 Is a Novel Regulator of Intracellular Calcium Homeostasis and Is Required for Spermatogenesis. PLoS ONE 2015, 10, e0127480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markin, A.; Khotina, V.; Zabudskaya, X.; Bogatyreva, A.; Starodubova, A.; Ivanova, E.; Nikiforov, N.; Orekhov, A. Disturbance of Mitochondrial Dynamics and Mitochondrial Therapies in Atherosclerosis. Life 2021, 11, 165. [Google Scholar] [CrossRef] [PubMed]
- Landowski, M.; Grindel, S.; Shahi, P.K.; Johnson, A.; Western, D.; Race, A.; Shi, F.; Benson, J.; Gao, M.; Santoirre, E.; et al. Modulation of Tmem135 Leads to Retinal Pigmented Epithelium Pathologies in Mice. Investig. Opthalmology Vis. Sci. 2020, 61, 16. [Google Scholar] [CrossRef]
- Spurthi, K.M.; Sarikhani, M.; Mishra, S.; Desingu, P.A.; Yadav, S.; Rao, S.; Maity, S.; Tamta, A.K.; Kumar, S.; Majumdar, S.; et al. Toll-like receptor 2 deficiency hyperactivates the FoxO1 transcription factor and induces aging-associated cardiac dysfunction in mice. J. Biol. Chem. 2018, 293, 13073–13089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, K.; Chen, D.; Riddle, D.L. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 2004, 131, 3897–3906. [Google Scholar] [CrossRef] [Green Version]
- Vellai, T.; Takacs-Vellai, K.; Zhang, Y.; Kovacs, A.L.; Orosz, L.; Müller, F. Influence of TOR kinase on lifespan in C. elegans. Nat. Cell Biol. 2003, 426, 620. [Google Scholar] [CrossRef] [PubMed]
- Kondo, M.; Yanase, S.; Ishii, T.; Hartman, P.S.; Matsumoto, K.; Ishii, N. The p38 signal transduction pathway participates in the oxidative stress-mediated translocation of DAF-16 to C. elegans nuclei. Mech. Ageing Dev. 2005, 126, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Fusakio, M.E.; Willy, J.A.; Wang, Y.; Mirek, E.T.; Al Baghdadi, R.J.T.; Adams, C.; Anthony, T.G.; Wek, R.C. Transcription factor ATF4 directs basal and stress-induced gene expression in the unfolded protein response and cholesterol metabolism in the liver. Mol. Biol. Cell 2016, 27, 1536–1551. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [Green Version]
- Khavandgar, Z.; Murshed, M. Sphingolipid metabolism and its role in the skeletal tissues. Cell. Mol. Life Sci. 2014, 72, 959–969. [Google Scholar] [CrossRef]
- Pulli, I.; Asghar, M.Y.; Kemppainen, K.; Törnquist, K. Sphingolipid-mediated calcium signaling and its pathological effects. Biochim. Biophys. Acta (BBA)—Bioenerg. 2018, 1865, 1668–1677. [Google Scholar] [CrossRef]
- Marchesini, N.; Osta, W.; Bielawski, J.; Luberto, C.; Obeid, L.M.; Hannun, Y.A. Role for Mammalian Neutral Sphingomyelinase 2 in Confluence-induced Growth Arrest of MCF7 Cells. J. Biol. Chem. 2004, 279, 25101–25111. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, K.; Tomiuk, S.; Wolff, G.; Stoffel, W. Cloning and characterization of the mammalian brain-specific, Mg2+-dependent neutral sphingomyelinase. Proc. Natl. Acad. Sci. USA 2000, 97, 5895–5900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miura, Y.; Gotoh, E.; Nara, F.; Nishijima, M.; Hanada, K. Hydrolysis of sphingosylphosphocholine by neutral sphingomyelinases. FEBS Lett. 2004, 557, 288–292. [Google Scholar] [CrossRef] [Green Version]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef] [PubMed]
- Airola, M.; Shanbhogue, P.; Shamseddine, A.A.; Guja, K.E.; Senkal, C.E.; Maini, R.; Bartke, N.; Wu, B.X.; Obeid, L.M.; Garcia-Diaz, M.; et al. Structure of human nSMase2 reveals an interdomain allosteric activation mechanism for ceramide generation. Proc. Natl. Acad. Sci. USA 2017, 114, E5549–E5558. [Google Scholar] [CrossRef] [Green Version]
- Natrajan, R.; Mackay, A.; Lambros, M.B.; Weigelt, B.; Wilkerson, P.M.; Manie, E.; Grigoriadis, A.; A’Hern, R.; Van Der Groep, P.; Kozarewa, I.; et al. A whole-genome massively parallel sequencing analysis of BRCA1 mutant oestrogen receptor-negative and -positive breast cancers. J. Pathol. 2012, 227, 29–41. [Google Scholar] [CrossRef]
- Li, J.; Zhong, H.-Y.; Zhang, Y.; Xiao, L.; Bai, L.-H.; Liu, S.-F.; Zhou, G.-B.; Zhang, G.-S. GTF2I-RARAis a novel fusion transcript in a t(7;17) variant of acute promyelocytic leukaemia with clinical resistance to retinoic acid. Br. J. Haematol. 2014, 168, 904–908. [Google Scholar] [CrossRef]
- Yu, Y.-P.; Liu, P.; Nelson, J.; Hamilton, R.L.; Bhargava, R.; Michalopoulos, G.; Chen, Q.; Zhang, J.; Ma, D.; Pennathur, A.; et al. Identification of recurrent fusion genes across multiple cancer types. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Franić, S.; Groen-Blokhuis, M.M.; Dolan, C.V.; Kattenberg, M.V.; Pool, R.; Xiao, X.; Scheet, P.A.; Ehli, E.A.; Davies, G.E.; van der Sluis, S.; et al. Intelligence: Shared genetic basis between Mendelian disorders and a polygenic trait. Eur. J. Hum. Genet. 2015, 23, 1378–1383. [Google Scholar] [CrossRef] [Green Version]
- Schweppe, D.K.; Huttlin, E.; Harper, J.; Gygi, S.P. BioPlex Display: An Interactive Suite for Large-Scale AP–MS Protein–Protein Interaction Data. J. Proteome Res. 2018, 17, 722–726. [Google Scholar] [CrossRef]
- Huttlin, E.L.; Bruckner, R.J.; Paulo, J.A.; Cannon, J.R.; Ting, L.; Baltier, K.; Colby, G.; Gebreab, F.; Gygi, M.P.; Parzen, H.; et al. Architecture of the human interactome defines protein communities and disease networks. Nature 2017, 545, 505–509. [Google Scholar] [CrossRef]
- Huttlin, E.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [Green Version]
- Aguayo-Orozco, A.; Bois, F.Y.; Brunak, S.; Taboureau, O. Analysis of Time-Series Gene Expression Data to Explore Mechanisms of Chemical-Induced Hepatic Steatosis Toxicity. Front. Genet. 2018, 9, 396. [Google Scholar] [CrossRef]
- Yang, W.; Li, Y.; He, F.; Wu, H. Microarray profiling of long non-coding RNA (lncRNA) associated with hypertrophic cardiomyopathy. BMC Cardiovasc. Disord. 2015, 15, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgkin, A.L.; Huxley, A.F. The components of membrane conductance in the giant axon of Loligo. J. Physiol. 1952, 116, 473–496. [Google Scholar] [CrossRef] [PubMed]
- Kadir, L.A.; Stacey, M.; Barrett-Jolley, R. Emerging Roles of the Membrane Potential: Action Beyond the Action Potential. Front. Physiol. 2018, 9, 1661. [Google Scholar] [CrossRef] [PubMed]






| Predicted Location | Major Expression in Tsue | Known Interactions with Organelles/Vesicles | Physiological Role | Implication in Human Disease |
|---|---|---|---|---|
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beasley, H.K.; Rodman, T.A.; Collins, G.V.; Hinton, A., Jr.; Exil, V. TMEM135 is a Novel Regulator of Mitochondrial Dynamics and Physiology with Implications for Human Health Conditions. Cells 2021, 10, 1750. https://doi.org/10.3390/cells10071750
Beasley HK, Rodman TA, Collins GV, Hinton A Jr., Exil V. TMEM135 is a Novel Regulator of Mitochondrial Dynamics and Physiology with Implications for Human Health Conditions. Cells. 2021; 10(7):1750. https://doi.org/10.3390/cells10071750
Chicago/Turabian StyleBeasley, Heather K., Taylor A. Rodman, Greg V. Collins, Antentor Hinton, Jr., and Vernat Exil. 2021. "TMEM135 is a Novel Regulator of Mitochondrial Dynamics and Physiology with Implications for Human Health Conditions" Cells 10, no. 7: 1750. https://doi.org/10.3390/cells10071750
APA StyleBeasley, H. K., Rodman, T. A., Collins, G. V., Hinton, A., Jr., & Exil, V. (2021). TMEM135 is a Novel Regulator of Mitochondrial Dynamics and Physiology with Implications for Human Health Conditions. Cells, 10(7), 1750. https://doi.org/10.3390/cells10071750