Cytokine-Mediated Regulation of ARG1 in Macrophages and Its Impact on the Control of Salmonella enterica Serovar Typhimurium Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Bone Marrow-Derived Macrophages and Culture Conditions
2.2. In Vitro Infection
2.3. Animals
2.4. In Vivo Infection
2.5. Pharmacological Inhibition of ARG1 In Vivo
2.6. Colony Forming Units
2.7. RNA Isolation, Reverse Transcription and TaqMan Quantitative Real-Time PCR (qRT-PCR)
2.8. Immunoblotting
2.9. Flow Cytometry
2.10. Detection of Interleukins and Nitrite
2.11. Urea Assay
2.12. Arginase 1 Activity Assay
2.13. Statistical Analysis
3. Results
3.1. IL-4-Dependent Arg1 Upregulation Is Ameliorated upon S.tm Infection
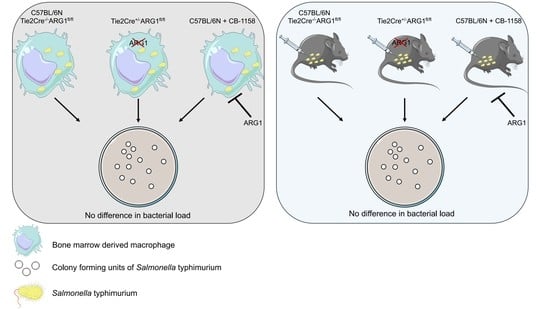
3.2. ARG1-Deficient BMDM from Tie2Cre+/−ARG1fl/fl Mice Did Not Show an Improved Control of S.tm Infection
3.3. ARG1-Deficient Tie2Cre+/−ARG1fl/fl Mice Showed Similar S.tm Control In Vivo as Compared to Wild-Type Littermates
3.4. Pharmacological Inhibition of ARG1 with CB-1158 Did Not Affect the Control of Systemic Salmonella Infection in Mice
3.5. ARG1 Is Dispensable for the Control of Salmonella by Macrophages Irrespective of the Presence or Absence of Functional NRAMP1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mills, C.D. M1 and M2 Macrophages: Oracles of Health and Disease. Crit. Rev. Immunol. 2012, 32, 463–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef]
- Gordon, S.; Martinez-Pomares, L. Physiological roles of macrophages. Pflug. Arch. 2017, 469, 365–374. [Google Scholar] [CrossRef] [Green Version]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef]
- Hannemann, N.; Cao, S.; Eriksson, D.; Schnelzer, A.; Jordan, J.; Eberhardt, M.; Schleicher, U.; Rech, J.; Ramming, A.; Uebe, S.; et al. Transcription factor Fra-1 targets arginase-1 to enhance macrophage-mediated inflammation in arthritis. J. Clin. Investig. 2019, 129, 2669–2684. [Google Scholar] [CrossRef] [Green Version]
- Weiss, G.; Schaible, U.E. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015, 264, 182–203. [Google Scholar] [CrossRef] [Green Version]
- Nathan, C.; Xie, Q.W. Nitric oxide synthases: Roles, tolls, and controls. Cell 1994, 78, 915–918. [Google Scholar] [CrossRef]
- Nairz, M.; Schleicher, U.; Schroll, A.; Sonnweber, T.; Theurl, I.; Ludwiczek, S.; Talasz, H.; Brandacher, G.; Moser, P.L.; Muckenthaler, M.U.; et al. Nitric oxide-mediated regulation of ferroportin-1 controls macrophage iron homeostasis and immune function in Salmonella infection. J. Exp. Med. 2013, 210, 855–873. [Google Scholar] [CrossRef] [PubMed]
- Bogdan, C. Nitric oxide synthase in innate and adaptive immunity: An update. Trends Immunol. 2015, 36, 161–178. [Google Scholar] [CrossRef] [PubMed]
- Schleicher, U.; Paduch, K.; Debus, A.; Obermeyer, S.; Konig, T.; Kling, J.C.; Ribechini, E.; Dudziak, D.; Mougiakakos, D.; Murray, P.J.; et al. TNF-Mediated Restriction of Arginase 1 Expression in Myeloid Cells Triggers Type 2 NO Synthase Activity at the Site of Infection. Cell Rep. 2016, 15, 1062–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piccolo, V.; Curina, A.; Genua, M.; Ghisletti, S.; Simonatto, M.; Sabo, A.; Amati, B.; Ostuni, R.; Natoli, G. Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat. Immunol. 2017, 18, 530–540. [Google Scholar] [CrossRef] [Green Version]
- Ostuni, R.; Natoli, G. Transcriptional control of macrophage diversity and specialization. Eur. J. Immunol. 2011, 41, 2486–2490. [Google Scholar] [CrossRef]
- El Kasmi, K.C.; Qualls, J.E.; Pesce, J.T.; Smith, A.M.; Thompson, R.W.; Henao-Tamayo, M.; Basaraba, R.J.; Konig, T.; Schleicher, U.; Koo, M.S.; et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat. Immunol. 2008, 9, 1399–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Muylder, G.; Daulouede, S.; Lecordier, L.; Uzureau, P.; Morias, Y.; Van Den Abbeele, J.; Caljon, G.; Herin, M.; Holzmuller, P.; Semballa, S.; et al. A Trypanosoma brucei kinesin heavy chain promotes parasite growth by triggering host arginase activity. PLoS Pathog. 2013, 9, e1003731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iniesta, V.; Carcelen, J.; Molano, I.; Peixoto, P.M.; Redondo, E.; Parra, P.; Mangas, M.; Monroy, I.; Campo, M.L.; Nieto, C.G.; et al. Arginase I induction during Leishmania major infection mediates the development of disease. Infect. Immun. 2005, 73, 6085–6090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paduch, K.; Debus, A.; Rai, B.; Schleicher, U.; Bogdan, C. Resolution of Cutaneous Leishmaniasis and Persistence of Leishmania major in the Absence of Arginase 1. J. Immunol. 2019, 202, 1453–1464. [Google Scholar] [CrossRef] [Green Version]
- Das, P.; Lahiri, A.; Lahiri, A.; Chakravortty, D. Modulation of the arginase pathway in the context of microbial pathogenesis: A metabolic enzyme moonlighting as an immune modulator. PLoS Pathog. 2010, 6, e1000899. [Google Scholar] [CrossRef] [Green Version]
- Wessling-Resnick, M. Nramp1 and Other Transporters Involved in Metal Withholding during Infection. J. Biol. Chem. 2015, 290, 18984–18990. [Google Scholar] [CrossRef] [Green Version]
- Haschka, D.; Hoffmann, A.; Weiss, G. Iron in immune cell function and host defense. Semin Cell Dev. Biol. 2020. [Google Scholar] [CrossRef]
- Cellier, M.F.; Courville, P.; Campion, C. Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect. 2007, 9, 1662–1670. [Google Scholar] [CrossRef]
- Vidal, S.; Tremblay, M.L.; Govoni, G.; Gauthier, S.; Sebastiani, G.; Malo, D.; Skamene, E.; Olivier, M.; Jothy, S.; Gros, P. The Ity/Lsh/Bcg locus: Natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J. Exp. Med. 1995, 182, 655–666. [Google Scholar] [CrossRef]
- Fritsche, G.; Nairz, M.; Libby, S.J.; Fang, F.C.; Weiss, G. Slc11a1 (Nramp1) impairs growth of Salmonella enterica serovar typhimurium in macrophages via stimulation of lipocalin-2 expression. J. Leukoc. Biol. 2012, 92, 353–359. [Google Scholar] [CrossRef] [Green Version]
- Fritsche, G.; Nairz, M.; Werner, E.R.; Barton, H.C.; Weiss, G. Nramp1-functionality increases iNOS expression via repression of IL-10 formation. Eur. J. Immunol. 2008, 38, 3060–3067. [Google Scholar] [CrossRef]
- Fritsche, G.; Dlaska, M.; Barton, H.; Theurl, I.; Garimorth, K.; Weiss, G. Nramp1 functionality increases inducible nitric oxide synthase transcription via stimulation of IFN regulatory factor 1 expression. J. Immunol. 2003, 171, 1994–1998. [Google Scholar] [CrossRef]
- Valdez, Y.; Diehl, G.E.; Vallance, B.A.; Grassl, G.A.; Guttman, J.A.; Brown, N.F.; Rosenberger, C.M.; Littman, D.R.; Gros, P.; Finlay, B.B. Nramp1 expression by dendritic cells modulates inflammatory responses during Salmonella Typhimurium infection. Cell Microbiol. 2008, 10, 1646–1661. [Google Scholar] [CrossRef] [Green Version]
- Bhutta, Z.A.; Gaffey, M.F.; Crump, J.A.; Steele, D.; Breiman, R.F.; Mintz, E.D.; Black, R.E.; Luby, S.P.; Levine, M.M. Typhoid Fever: Way Forward. Am. J. Trop. Med. Hyg. 2018, 99, 89–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumler, A.; Fang, F.C. Host specificity of bacterial pathogens. Cold Spring Harb. Perspect Med. 2013, 3, a010041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchmeier, N.A.; Heffron, F. Inhibition of macrophage phagosome-lysosome fusion by Salmonella typhimurium. Infect. Immun. 1991, 59, 2232–2238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fields, P.I.; Swanson, R.V.; Haidaris, C.G.; Heffron, F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 1986, 83, 5189–5193. [Google Scholar] [CrossRef] [Green Version]
- Chakravortty, D.; Hansen-Wester, I.; Hensel, M. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J. Exp. Med. 2002, 195, 1155–1166. [Google Scholar] [CrossRef]
- Lahiri, A.; Lahiri, A.; Iyer, N.; Das, P.; Chakravortty, D. Visiting the cell biology of Salmonella infection. Microbes Infect. 2010, 12, 809–818. [Google Scholar] [CrossRef]
- Mastroeni, P.; Grant, A.J. Spread of Salmonella enterica in the body during systemic infection: Unravelling host and pathogen determinants. Expert Rev. Mol. Med. 2011, 13, e12. [Google Scholar] [CrossRef] [PubMed]
- Navarre, W.W.; Zou, S.B.; Roy, H.; Xie, J.L.; Savchenko, A.; Singer, A.; Edvokimova, E.; Prost, L.R.; Kumar, R.; Ibba, M.; et al. PoxA, yjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol. Cell 2010, 39, 209–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dichtl, S.; Demetz, E.; Haschka, D.; Tymoszuk, P.; Petzer, V.; Nairz, M.; Seifert, M.; Hoffmann, A.; Brigo, N.; Wurzner, R.; et al. Dopamine Is a Siderophore-Like Iron Chelator That Promotes Salmonella enterica Serovar Typhimurium Virulence in Mice. mBio 2019, 10, e02624-18. [Google Scholar] [CrossRef] [Green Version]
- Sin, Y.Y.; Ballantyne, L.L.; Mukherjee, K.; St Amand, T.; Kyriakopoulou, L.; Schulze, A.; Funk, C.D. Inducible arginase 1 deficiency in mice leads to hyperargininemia and altered amino acid metabolism. PLoS ONE 2013, 8, e80001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nairz, M.; Ferring-Appel, D.; Casarrubea, D.; Sonnweber, T.; Viatte, L.; Schroll, A.; Haschka, D.; Fang, F.C.; Hentze, M.W.; Weiss, G.; et al. Iron Regulatory Proteins Mediate Host Resistance to Salmonella Infection. Cell Host Microbe 2015, 18, 254–261. [Google Scholar] [CrossRef] [Green Version]
- Steggerda, S.M.; Bennett, M.K.; Chen, J.; Emberley, E.; Huang, T.; Janes, J.R.; Li, W.; MacKinnon, A.L.; Makkouk, A.; Marguier, G.; et al. Inhibition of arginase by CB-1158 blocks myeloid cell-mediated immune suppression in the tumor microenvironment. J. Immunother. Cancer 2017, 5, 101. [Google Scholar] [CrossRef]
- Grobben, Y.; Uitdehaag, J.C.M.; Willemsen-Seegers, N.; Tabak, W.W.A.; de Man, J.; Buijsman, R.C.; Zaman, G.J.R. Structural insights into human Arginase-1 pH dependence and its inhibition by the small molecule inhibitor CB-1158. J. Struct. Biol. X 2020, 4, 100014. [Google Scholar] [CrossRef]
- Ludwiczek, S.; Theurl, I.; Muckenthaler, M.U.; Jakab, M.; Mair, S.M.; Theurl, M.; Kiss, J.; Paulmichl, M.; Hentze, M.W.; Ritter, M.; et al. Ca2+ channel blockers reverse iron overload by a new mechanism via divalent metal transporter-1. Nat. Med. 2007, 13, 448–454. [Google Scholar] [CrossRef]
- Theurl, I.; Hilgendorf, I.; Nairz, M.; Tymoszuk, P.; Haschka, D.; Asshoff, M.; He, S.; Gerhardt, L.M.; Holderried, T.A.; Seifert, M.; et al. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat. Med. 2016, 22, 945–951. [Google Scholar] [CrossRef]
- Nairz, M.; Theurl, I.; Schroll, A.; Theurl, M.; Fritsche, G.; Lindner, E.; Seifert, M.; Crouch, M.L.; Hantke, K.; Akira, S.; et al. Absence of functional Hfe protects mice from invasive Salmonella enterica serovar Typhimurium infection via induction of lipocalin-2. Blood 2009, 114, 3642–3651. [Google Scholar] [CrossRef] [Green Version]
- Corraliza, I.M.; Campo, M.L.; Soler, G.; Modolell, M. Determination of arginase activity in macrophages: A micromethod. J. Immunol. Methods 1994, 174, 231–235. [Google Scholar] [CrossRef]
- Iyer, R.K.; Yoo, P.K.; Kern, R.M.; Rozengurt, N.; Tsoa, R.; O’Brien, W.E.; Yu, H.; Grody, W.W.; Cederbaum, S.D. Mouse model for human arginase deficiency. Mol. Cell Biol. 2002, 22, 4491–4498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerrinnes, T.; Winter, M.G.; Young, B.M.; Diaz-Ochoa, V.E.; Winter, S.E.; Tsolis, R.M. Utilization of Host Polyamines in Alternatively Activated Macrophages Promotes Chronic Infection by Brucella abortus. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldman-Pinkovich, A.; Balno, C.; Strasser, R.; Zeituni-Molad, M.; Bendelak, K.; Rentsch, D.; Ephros, M.; Wiese, M.; Jardim, A.; Myler, P.J.; et al. An Arginine Deprivation Response Pathway Is Induced in Leishmania during Macrophage Invasion. PLoS Pathog. 2016, 12, e1005494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Telser, J.; Volani, C.; Hilbe, R.; Seifert, M.; Brigo, N.; Paglia, G.; Weiss, G. Metabolic reprogramming of Salmonella infected macrophages and its modulation by iron availability and the mTOR pathway. Microb. Cell 2019, 6, 531–543. [Google Scholar] [CrossRef]
- Cuervo, H.; Pineda, M.A.; Aoki, M.P.; Gea, S.; Fresno, M.; Girones, N. Inducible nitric oxide synthase and arginase expression in heart tissue during acute Trypanosoma cruzi infection in mice: Arginase I is expressed in infiltrating CD68+ macrophages. J. Infect. Dis. 2008, 197, 1772–1782. [Google Scholar] [CrossRef] [Green Version]
- Lahiri, A.; Das, P.; Chakravortty, D. Arginase modulates Salmonella induced nitric oxide production in RAW264.7 macrophages and is required for Salmonella pathogenesis in mice model of infection. Microbes Infect. 2008, 10, 1166–1174. [Google Scholar] [CrossRef]
- Gumy, A.; Louis, J.A.; Launois, P. The murine model of infection with Leishmania major and its importance for the deciphering of mechanisms underlying differences in Th cell differentiation in mice from different genetic backgrounds. Int. J. Parasitol. 2004, 34, 433–444. [Google Scholar] [CrossRef]
- Gerichter, C.B. The dissemination of Salmonella typhi, S. paratyphi A and S. paratyphi B through the organs of the white mouse by oral infection. J. Hyg. 1960, 58, 307–319. [Google Scholar] [CrossRef] [Green Version]
- Higginson, E.E.; Simon, R.; Tennant, S.M. Animal Models for Salmonellosis: Applications in Vaccine Research. Clin. Vaccine Immunol. 2016, 23, 746–756. [Google Scholar] [CrossRef] [Green Version]
- Mathur, R.; Oh, H.; Zhang, D.; Park, S.G.; Seo, J.; Koblansky, A.; Hayden, M.S.; Ghosh, S. A mouse model of Salmonella typhi infection. Cell 2012, 151, 590–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, A.D.; Slauch, J.M. Mechanisms of Salmonella pathogenesis in animal models. Hum. Ecol. Risk Assess 2017, 23, 1877–1892. [Google Scholar] [CrossRef] [PubMed]
- Mastroeni, P.; Vazquez-Torres, A.; Fang, F.C.; Xu, Y.; Khan, S.; Hormaeche, C.E.; Dougan, G. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 2000, 192, 237–248. [Google Scholar] [CrossRef]
- Popovic, P.J.; Zeh, H.J., 3rd; Ochoa, J.B. Arginine and immunity. J. Nutr. 2007, 137, 1681S–1686S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rath, M.; Muller, I.; Kropf, P.; Closs, E.I.; Munder, M. Metabolism via Arginase or Nitric Oxide Synthase: Two Competing Arginine Pathways in Macrophages. Front. Immunol. 2014, 5, 532. [Google Scholar] [CrossRef] [Green Version]
- Gogoi, M.; Datey, A.; Wilson, K.T.; Chakravortty, D. Dual role of arginine metabolism in establishing pathogenesis. Curr. Opin. Microbiol. 2016, 29, 43–48. [Google Scholar] [CrossRef] [Green Version]







| Murine Gene | Forward Primer 5′-3′ | Reverse Primer 5′-3′ | Probe |
|---|---|---|---|
| Hprt | GACCGGTCCCGTCATGC | TCATAACCTGGTTCATCATCGC | ACCCGCAGTCCCAGCGTCGTC |
| Arg1 | AACACGGCAGTGGCTTTAAC | GAGGAGAAGGCGTTTGCTTA | TGGCTTATGGTTACCCTCCCGTTG |
| iNos | CAGCTGGGCTGTACAAACCTT | CATTGGAAGTGAAGCGTTTCG | CGG GCA GCC TGT GAG ACC TTT GA |
| Tnf | TTCTATGGCCCAGACCCTCA | TTGCTACGACGTGGGCTACA | CTCAGATCATCTTCTCAAAATTCGAGTGACAAGC |
| Il-6 | TGTTCTCTGGGAAATCGTGGA | AAGTGCATCATCGTTGTTCATACA | ATGAGAAAAGAGTTGTGCAATGGCAATTCTG |
| Il-10 | CCAGAGCCACATGCTCCTAGA | TGGTCCTTTGTTTGAAAGAAAGTCT | TGCGGACTGCCTTCAGCCAGG |
| Murine Gene | TaqMan® Genexpressionassays |
|---|---|
| Hprt | Mm00446968_m1 |
| Mrc1 | Mm00485148_m1 |
| Chil3 | Mm00657889_mH |
| Il-6 | Mm00446190_m1 |
| Tnf | Mm00443258_m1 |
| Detected Protein | Antibody Dilution | Commercial Source | Catalogue Number |
|---|---|---|---|
| ARG1 | 1:2000 | Novusbio (Littelton, CO, USA) | NBP1-32731 |
| iNOS | 1:1000 | Abcam | ab3523 |
| β-ACTIN | 1:500 | Sigma-Aldrich | A 2066 |
| Antibody against | Conjugated Fluorochrome | Commercial Source | Catalogue Number |
|---|---|---|---|
| CD3 | PE-eFluor 610 | Invitrogen | 61-0031-82 |
| CD19 | PE-eFluor 610 | Invitrogen | 61-193-82 |
| CD49b | PE-CF594 | BD Horizon | 562453 |
| CD11b | BB515 | BD Horizon | 564454 |
| CD45 | APC-R700 | BD Horizon | 565478 |
| F4/80 | BV421 | BD Horizon | 565411 |
| Ly6C | BV510 | BD Horizon | 563040 |
| Ly6G | Percp-eflour 710 | Invitrogen | 46-9668-82 |
| iNOS | PeCy7 | Invitrogen | 25-5920-80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brigo, N.; Pfeifhofer-Obermair, C.; Tymoszuk, P.; Demetz, E.; Engl, S.; Barros-Pinkelnig, M.; Dichtl, S.; Fischer, C.; Valente De Souza, L.; Petzer, V.; et al. Cytokine-Mediated Regulation of ARG1 in Macrophages and Its Impact on the Control of Salmonella enterica Serovar Typhimurium Infection. Cells 2021, 10, 1823. https://doi.org/10.3390/cells10071823
Brigo N, Pfeifhofer-Obermair C, Tymoszuk P, Demetz E, Engl S, Barros-Pinkelnig M, Dichtl S, Fischer C, Valente De Souza L, Petzer V, et al. Cytokine-Mediated Regulation of ARG1 in Macrophages and Its Impact on the Control of Salmonella enterica Serovar Typhimurium Infection. Cells. 2021; 10(7):1823. https://doi.org/10.3390/cells10071823
Chicago/Turabian StyleBrigo, Natascha, Christa Pfeifhofer-Obermair, Piotr Tymoszuk, Egon Demetz, Sabine Engl, Marina Barros-Pinkelnig, Stefanie Dichtl, Christine Fischer, Lara Valente De Souza, Verena Petzer, and et al. 2021. "Cytokine-Mediated Regulation of ARG1 in Macrophages and Its Impact on the Control of Salmonella enterica Serovar Typhimurium Infection" Cells 10, no. 7: 1823. https://doi.org/10.3390/cells10071823
APA StyleBrigo, N., Pfeifhofer-Obermair, C., Tymoszuk, P., Demetz, E., Engl, S., Barros-Pinkelnig, M., Dichtl, S., Fischer, C., Valente De Souza, L., Petzer, V., von Raffay, L., Hilbe, R., Berger, S., Seifert, M., Schleicher, U., Bogdan, C., & Weiss, G. (2021). Cytokine-Mediated Regulation of ARG1 in Macrophages and Its Impact on the Control of Salmonella enterica Serovar Typhimurium Infection. Cells, 10(7), 1823. https://doi.org/10.3390/cells10071823