Anti-Human CD9 Fab Fragment Antibody Blocks the Extracellular Vesicle-Mediated Increase in Malignancy of Colon Cancer Cells
Abstract
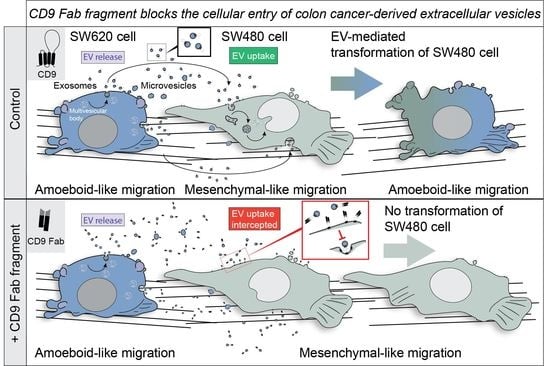
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Lentiviral Infection
2.3. Plasmid and Transfection
2.4. Isolation and Characterization of EVs
2.5. CD9 Antibody Fab Fragment
2.6. Immunoblotting
2.7. Cell–EV Incubation
2.8. Confocal Laser Scanning Microscopy
2.9. Stochastic Optical Reconstruction Microscopy
2.10. Flow Cytometry
2.11. Cell Migration
2.12. Statistical Analysis
3. Results
3.1. Characterization of EVs Released by SW620 Cells
3.2. Internalization of SW620 Cell-Derived CD9-GFP+ EVs into SW480 Cells: Impact of CD9 Ab
3.3. Pro-Metastatic Morphological Alterations of SW480 Cells by SW620 Cell-Derived EVs Are Blocked by CD9 Fab
3.4. SW620 Cell-Derived EV-Induced Migration of SW480 Cells Is Impeded by CD9 Fab
3.5. CD9 Is Essential for Pro-Metastatic Morphological Alterations of SW480 Cells Mediated by SW620 Cell-Derived EVs
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, K.; Wang, Z.; Hackert, T.; Pitzer, C.; Zöller, M. Tspan8 and Tspan8/CD151 knockout mice unravel the contribution of tumor and host exosomes to tumor progression. J. Exp. Clin. Cancer Res. 2018, 37, 312. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Cocucci, E.; Racchetti, G.; Meldolesi, J. Shedding microvesicles: Artefacts no more. Trends Cell Biol. 2009, 19, 43–51. [Google Scholar] [CrossRef]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef]
- Piper, R.C.; Katzmann, D.J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007, 23, 519–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Ratajczak, J.; Wysoczynski, M.; Hayek, F.; Janowska-Wieczorek, A.; Ratajczak, M.Z. Membrane-derived microvesicles: Important and underappreciated mediators of cell-to-cell communication. Leukemia 2006, 20, 1487–1495. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Schillaci, O.; Fontana, S.; Monteleone, F.; Taverna, S.; Di Bella, M.A.; Di Vizio, D.; Alessandro, R. Exosomes from metastatic cancer cells transfer amoeboid phenotype to non-metastatic cells and increase endothelial permeability: Their emerging role in tumor heterogeneity. Sci. Rep. 2017, 7, 4711. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Stolz, D.B.; Sullivan, M.L.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [Green Version]
- Corbeil, D.; Santos, M.F.; Karbanová, J.; Kurth, T.; Rappa, G.; Lorico, A. Uptake and Fate of Extracellular Membrane Vesicles: Nucleoplasmic Reticulum-Associated Late Endosomes as a New Gate to Intercellular Communication. Cells 2020, 9, 1931. [Google Scholar] [CrossRef]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osti, D.; Del Bene, M.; Rappa, G.; Santos, M.; Matafora, V.; Richichi, C.; Faletti, S.; Beznoussenko, G.V.; Mironov, A.; Bachi, A.; et al. Clinical Significance of Extracellular Vesicles in Plasma from Glioblastoma Patients. Clin. Cancer Res. 2019, 25, 266–276. [Google Scholar] [CrossRef] [Green Version]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [Green Version]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell 2019, 49, 347–360. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef] [Green Version]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobrie, A.; Krumeich, S.; Reyal, F.; Recchi, C.; Moita, L.F.; Seabra, M.C.; Ostrowski, M.; Théry, C. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012, 72, 4920–4930. [Google Scholar] [CrossRef] [Green Version]
- Bobrie, A.; Colombo, M.; Krumeich, S.; Raposo, G.; Théry, C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles 2012, 1, 18397. [Google Scholar] [CrossRef] [PubMed]
- Savina, A.; Furlan, M.; Vidal, M.; Colombo, M.I. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 2003, 278, 20083–20090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalmin, F.; Ladoire, S.; Mignot, G.; Vincent, J.; Bruchard, M.; Remy-Martin, J.P.; Boireau, W.; Rouleau, A.; Simon, B.; Lanneau, D.; et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Investig. 2010, 120, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Datta, A.; Kim, H.; McGee, L.; Johnson, A.E.; Talwar, S.; Marugan, J.; Southall, N.; Hu, X.; Lal, M.; Mondal, D.; et al. High-throughput screening identified selective inhibitors of exosome biogenesis and secretion: A drug repurposing strategy for advanced cancer. Sci. Rep. 2018, 8, 8161. [Google Scholar] [CrossRef] [Green Version]
- McAndrews, K.M.; Kalluri, R. Mechanisms associated with biogenesis of exosomes in cancer. Mol. Cancer 2019, 18, 52. [Google Scholar] [CrossRef]
- Kogure, A.; Yoshioka, Y.; Ochiya, T. Extracellular Vesicles in Cancer Metastasis: Potential as Therapeutic Targets and Materials. Int. J. Mol. Sci. 2020, 21, 4463. [Google Scholar] [CrossRef]
- Nishida-Aoki, N.; Tominaga, N.; Takeshita, F.; Sonoda, H.; Yoshioka, Y.; Ochiya, T. Disruption of Circulating Extracellular Vesicles as a Novel Therapeutic Strategy against Cancer Metastasis. Mol. Ther. 2017, 25, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Kato, T.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles 2013, 2, 20424. [Google Scholar] [CrossRef]
- Reyes, R.; Cardeñes, B.; Machado-Pineda, Y.; Cabañas, C. Tetraspanin CD9: A Key Regulator of Cell Adhesion in the Immune System. Front. Immunol. 2018, 9, 863. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, C.; Colas, L.; Magnan, A.; Brouard, S. CD9 Tetraspanin: A New Pathway for the Regulation of Inflammation? Front. Immunol. 2018, 9, 2316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorico, A.; Lorico-Rappa, M.; Karbanová, J.; Corbeil, D.; Pizzorno, G. CD9, a tetraspanin target for cancer therapy? Exp. Biol. Med. (Maywood) 2021, 9, 1121–1138. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [Green Version]
- Miki, Y.; Yashiro, M.; Okuno, T.; Kitayama, K.; Masuda, G.; Hirakawa, K.; Ohira, M. CD9-positive exosomes from cancer-associated fibroblasts stimulate the migration ability of scirrhous-type gastric cancer cells. Br. J. Cancer 2018, 118, 867–877. [Google Scholar] [CrossRef] [Green Version]
- Rappa, G.; Green, T.M.; Lorico, A. The nuclear pool of tetraspanin CD9 contributes to mitotic processes in human breast carcinoma. Mol. Cancer Res. 2014, 12, 1840–1850. [Google Scholar] [CrossRef] [Green Version]
- Rappa, G.; Santos, M.F.; Green, T.M.; Karbanová, J.; Hassler, J.; Bai, Y.; Barsky, S.H.; Corbeil, D.; Lorico, A. Nuclear transport of cancer extracellular vesicle-derived biomaterials through nuclear envelope invagination-associated late endosomes. Oncotarget 2017, 8, 14443–14461. [Google Scholar] [CrossRef] [Green Version]
- Le Naour, F.; Rubinstein, E.; Jasmin, C.; Prenant, M.; Boucheix, C. Severely reduced female fertility in CD9-deficient mice. Science 2000, 287, 319–321. [Google Scholar] [CrossRef]
- Miyado, K.; Yamada, G.; Yamada, S.; Hasuwa, H.; Nakamura, Y.; Ryu, F.; Suzuki, K.; Kosai, K.; Inoue, K.; Ogura, A.; et al. Requirement of CD9 on the egg plasma membrane for fertilization. Science 2000, 287, 321–324. [Google Scholar] [CrossRef]
- Miyado, K.; Yoshida, K.; Yamagata, K.; Sakakibara, K.; Okabe, M.; Wang, X.; Miyamoto, K.; Akutsu, H.; Kondo, T.; Takahashi, Y.; et al. The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc. Natl. Acad. Sci. USA 2008, 105, 12921–12926. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.S.; Tung, K.S.; Coonrod, S.A.; Takahashi, Y.; Bigler, D.; Chang, A.; Yamashita, Y.; Kincade, P.W.; Herr, J.C.; White, J.M. Role of the integrin-associated protein CD9 in binding between sperm ADAM 2 and the egg integrin alpha6beta1: Implications for murine fertilization. Proc. Natl. Acad. Sci. USA 1999, 96, 11830–11835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caballero, J.N.; Frenette, G.; Belleannée, C.; Sullivan, R. CD9-positive microvesicles mediate the transfer of molecules to Bovine Spermatozoa during epididymal maturation. PLoS ONE 2013, 8, e65364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaprielian, Z.; Cho, K.O.; Hadjiargyrou, M.; Patterson, P.H. CD9, a major platelet cell surface glycoprotein, is a ROCA antigen and is expressed in the nervous system. J. Neurosci. 1995, 15, 562–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, R.C.; Rubinstein, E.; Worthington, R.E.; Boucheix, C. Extensive C1q-complement initiated lysis of human platelets by IgG subclass murine monoclonal antibodies to the CD9 antigen. Thromb. Res. 1990, 59, 831–839. [Google Scholar] [CrossRef]
- Hato, T.; Ikeda, K.; Yasukawa, M.; Watanabe, A.; Kobayashi, Y. Exposure of platelet fibrinogen receptors by a monoclonal antibody to CD9 antigen. Blood 1988, 72, 224–229. [Google Scholar] [CrossRef] [Green Version]
- Worthington, R.E.; Carroll, R.C.; Boucheix, C. Platelet activation by CD9 monoclonal antibodies is mediated by the Fc gamma II receptor. Br. J. Haematol. 1990, 74, 216–222. [Google Scholar] [CrossRef]
- Jennings, L.K.; Fox, C.F.; Kouns, W.C.; McKay, C.P.; Ballou, L.R.; Schultz, H.E. The activation of human platelets mediated by anti-human platelet p24/CD9 monoclonal antibodies. J. Biol. Chem. 1990, 265, 3815–3822. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Suzuki, M.; Kido, H.; Sakane, H.; Hada, S.; Yamaguchi, K.; Fukuroi, T.; Yanabu, M.; Nagata, H.; Nomura, S.; et al. Antithrombotic effect of an anti-glycoprotein IIB/IIIA antibody in primate lethal thrombosis. Thromb. Res. 1993, 70, 245–254. [Google Scholar] [CrossRef]
- Kobayashi, H.; Hosono, O.; Iwata, S.; Kawasaki, H.; Kuwana, M.; Tanaka, H.; Dang, N.H.; Morimoto, C. The tetraspanin CD9 is preferentially expressed on the human CD4(+)CD45RA+ naive T cell population and is involved in T cell activation. Clin. Expl. Immunol. 2004, 137, 101–108. [Google Scholar] [CrossRef]
- Santos, M.F.; Rappa, G.; Karbanová, J.; Vanier, C.; Morimoto, C.; Corbeil, D.; Lorico, A. Anti-human CD9 antibody Fab fragment impairs the internalization of extracellular vesicles and the nuclear transfer of their cargo proteins. J. Cell Mol. Med. 2019, 23, 4408–4421. [Google Scholar] [CrossRef] [Green Version]
- Pang, H.; Liu, L.; Sun, X.; Xi, W.; Bao, Y.; Wu, L.; Shan, J.; Wang, Z.; Guo, Y.; Zhao, C. Exosomes derived from colon cancer cells and plasma of colon cancer patients promote migration of SW480 cells through Akt/mTOR pathway. Pathol. Res. Pract. 2021, 222, 153454. [Google Scholar] [CrossRef] [PubMed]
- Leibovitz, A.; Stinson, J.C.; McCombs, W.B., 3rd; McCoy, C.E.; Mazur, K.C.; Mabry, N.D. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976, 36, 4562–4569. [Google Scholar] [PubMed]
- Hewitt, R.E.; McMarlin, A.; Kleiner, D.; Wersto, R.; Martin, P.; Tsokos, M.; Stamp, G.W.; Stetler-Stevenson, W.G. Validation of a model of colon cancer progression. J. Pathol. 2000, 192, 446–454. [Google Scholar] [CrossRef]
- Melcher, R.; Steinlein, C.; Feichtinger, W.; Müller, C.R.; Menzel, T.; Lührs, H.; Scheppach, W.; Schmid, M. Spectral karyotyping of the human colon cancer cell lines SW480 and SW620. Cytogenet. Genome Res. 2000, 88, 145–152. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- EV-TRACT Consortium; Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, O.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar]
- Santos, M.F.; Rappa, G.; Karbanová, J.; Fontana, S.; Di Bella, M.A.; Pope, M.R.; Parrino, B.; Cascioferro, S.M.; Vistoli, G.; Diana, P.; et al. Itraconazole inhibits nuclear delivery of extracellular vesicle cargo by disrupting the entry of late endosomes into the nucleoplasmic reticulum. J. Extracell. Vesicles 2021, 10, e12132. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Qian, F.; Li, Y.P.; Sheng, X.; Zhang, Z.C.; Song, R.; Dong, W.; Cao, S.X.; Hua, Z.C.; Xu, Q. PRL-3 siRNA inhibits the metastasis of B16-BL6 mouse melanoma cells in vitro and in vivo. Mol. Med. 2007, 13, 151–159. [Google Scholar] [CrossRef]
- Cajot, J.F.; Sordat, I.; Silvestre, T.; Sordat, B. Differential display cloning identifies motility-related protein (MRP1/CD9) as highly expressed in primary compared to metastatic human colon carcinoma cells. Cancer Res. 1997, 57, 2593–2597. [Google Scholar]
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020, 219, e201904113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suwakulsiri, W.; Rai, A.; Xu, R.; Chen, M.; Greening, D.W.; Simpson, R.J. Proteomic profiling reveals key cancer progression modulators in shed microvesicles released from isogenic human primary and metastatic colorectal cancer cell lines. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 140171. [Google Scholar] [CrossRef] [PubMed]
- Popēna, I.; Ābols, A.; Saulīte, L.; Pleiko, K.; Zandberga, E.; Jēkabsons, K.; Endzelinš, E.; Llorente, A.; Linē, A.; Riekstina, U. Effect of colorectal cancer-derived extracellular vesicles on the immunophenotype and cytokine secretion profile of monocytes and macrophages. Cell Commun. Signal. 2018, 16, 17. [Google Scholar] [CrossRef] [Green Version]
- Depciuch, J.; Klebowski, B.; Stec, M.; Szatanek, R.; Weglarczyk, K.; Baj-Krzyworzeka, M.; Parlinska-Wojtan, M.; Baran, J. Similarities in the General Chemical Composition of Colon Cancer Cells and Their Microvesicles Investigated by Spectroscopic Methods-Potential Clinical Relevance. Int. J. Mol. Sci. 2020, 21, 1826. [Google Scholar] [CrossRef] [Green Version]
- Han, C.; Kang, H.; Yi, J.; Kang, M.; Lee, H.; Kwon, Y.; Jung, J.; Lee, J.; Park, J. Single-vesicle imaging and co-localization analysis for tetraspanin profiling of individual extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12047. [Google Scholar] [CrossRef]
- Santos, M.F.; Rappa, G.; Karbanová, J.; Kurth, T.; Corbeil, D.; Lorico, A. VAMP-associated protein-A and oxysterol-binding protein-related protein 3 promote the entry of late endosomes into the nucleoplasmic reticulum. J. Biol. Chem. 2018, 293, 13834–13848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fackler, O.T.; Grosse, R. Cell motility through plasma membrane blebbing. J. Cell Biol. 2008, 181, 879–884. [Google Scholar] [CrossRef] [Green Version]
- de Toledo, M.; Anguille, C.; Roger, L.; Roux, P.; Gadea, G. Cooperative anti-invasive effect of Cdc42/Rac1 activation and ROCK inhibition in SW620 colorectal cancer cells with elevated blebbing activity. PLoS ONE 2012, 7, e48344. [Google Scholar] [CrossRef] [Green Version]
- Ono, M.; Handa, K.; Sonnino, S.; Withers, D.A.; Nagai, H.; Hakomori, S. GM3 ganglioside inhibits CD9-facilitated haptotactic cell motility: Coexpression of GM3 and CD9 is essential in the downregulation of tumor cell motility and malignancy. Biochemistry 2001, 40, 6414–6421. [Google Scholar] [CrossRef]
- Ovalle, S.; Gutiérrez-López, M.D.; Olmo, N.; Turnay, J.; Lizarbe, M.A.; Majano, P.; Molina-Jimenez, F.; López-Cabrera, M.; Yáñez-Mó, M.; Sánchez-Madrid, F.; et al. The tetraspanin CD9 inhibits the proliferation and tumorigenicity of human colon carcinoma cells. Int. J. Cancer 2007, 121, 2140–2152. [Google Scholar] [CrossRef]
- Zöller, M. Tetraspanins: Push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 2009, 9, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Orchard-Webb, D.J.; Lee, T.C.; Cook, G.P.; Blair, G.E. CUB domain containing protein 1 (CDCP1) modulates adhesion and motility in colon cancer cells. BMC Cancer 2014, 14, 754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Zhong, T.; Zhong, J.; Tang, Y.; Ling, B.; Wang, L. MicroRNA129 inhibits colorectal cancer cell proliferation, invasion and epithelialtomesenchymal transition by targeting SOX4. Oncol. Rep. 2021, 45, 61. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Ma, L.; Shen, L.; Lei, Y.; Guo, L.; Deng, Y.; Ding, Y. MIF/SCL3A2 depletion inhibits the proliferation and metastasis of colorectal cancer cells via the AKT/GSK-3beta pathway and cell iron death. J. Cell Mol. Med. 2022, 26, 3410–3422. [Google Scholar] [CrossRef] [PubMed]
- Ji, R.; Zhang, B.; Zhang, X.; Xue, J.; Yuan, X.; Yan, Y.; Wang, M.; Zhu, W.; Qian, H.; Xu, W. Exosomes derived from human mesenchymal stem cells confer drug resistance in gastric cancer. Cell Cycle 2015, 14, 2473–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steeg, P.S. Targeting metastasis. Nat. Rev. Cancer 2016, 16, 201–218. [Google Scholar] [CrossRef]
- Beans, C. News Feature: Targeting metastasis to halt cancer’s spread. Proc. Natl. Acad. Sci. USA 2018, 115, 12539–12543. [Google Scholar] [CrossRef] [Green Version]
- Shafqat-Abbasi, H.; Kowalewski, J.M.; Kiss, A.; Gong, X.; Hernandez-Varas, P.; Berge, U.; Jafari-Mamaghani, M.; Lock, J.G.; Stromblad, S. An analysis toolbox to explore mesenchymal migration heterogeneity reveals adaptive switching between distinct modes. Elife 2016, 5, e11384. [Google Scholar] [CrossRef] [PubMed]
- Wolf, K.; Mazo, I.; Leung, H.; Engelke, K.; von Andrian, U.H.; Deryugina, E.I.; Strongin, A.Y.; Brocker, E.B.; Friedl, P. Compensation mechanism in tumor cell migration: Mesenchymal-amoeboid transition after blocking of pericellular proteolysis. J. Cell Biol. 2003, 160, 267–277. [Google Scholar] [CrossRef] [Green Version]
- Read, J.; Ingram, A.; Al Saleh, H.A.; Platko, K.; Gabriel, K.; Kapoor, A.; Pinthus, J.; Majeed, F.; Qureshi, T.; Al-Nedawi, K. Nuclear transportation of exogenous epidermal growth factor receptor and androgen receptor via extracellular vesicles. Eur. J. Cancer 2017, 70, 62–74. [Google Scholar] [CrossRef]
- Alfandari, D.; Ben Ami Pilo, H.; Abou Karam, P.; Dagan, O.; Joubran, C.; Rotkopf, R.; Regev-Rudzki, N.; Porat, Z. Monitoring Distribution Dynamics of EV RNA Cargo Within Recipient Monocytes and Macrophages. Front. Cell. Infect. Microbiol. 2021, 11, 739628. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, O.V.; Yang, X.; Kolesnikova, T.V.; Hemler, M.E. Evidence for specific tetraspanin homodimers: Inhibition of palmitoylation makes cysteine residues available for cross-linking. Biochem. J. 2004, 377, 407–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes, R.; Monjas, A.; Yánez-Mó, M.; Cardeñes, B.; Morlino, G.; Gilsanz, A.; Machado-Pineda, Y.; Lafuente, E.; Monk, P.; Sánchez-Madrid, F.; et al. Different states of integrin LFA-1 aggregation are controlled through its association with tetraspanin CD9. Biochim. Biophys. Acta 2015, 1853, 2464–2480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barreiro, O.; Yáñez-Mó, M.; Sala-Valdés, M.; Gutiérrez-Lopez, M.D.; Ovalle, S.; Higginbottom, A.; Monk, P.N.; Cabañas, C.; Sánchez-Madrid, F. Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood 2005, 105, 2852–2861. [Google Scholar] [CrossRef] [PubMed]
- Barreiro, O.; Zamai, M.; Yañez-Mó, M.; Tejera, E.; López-Romero, P.; Monk, P.N.; Gratton, E.; Caiolfa, V.R.; Sánchez-Madrid, F. Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J. Cell Biol. 2008, 183, 527–542. [Google Scholar] [CrossRef] [Green Version]
- Gilsanz, A.; Sánchez-Martín, L.; Gutiérrez-López, M.D.; Ovalle, S.; Machado-Pineda, Y.; Reyes, R.; Swart, G.W.; Figdor, C.G.; Lafuente, E.M.; Cabañas, C. ALCAM/CD166 adhesive function is regulated by the tetraspanin CD9. Cell Mol. Life Sci. 2013, 70, 475–493. [Google Scholar] [CrossRef]
- Rappa, G.; Green, T.M.; Karbanová, J.; Corbeil, D.; Lorico, A. Tetraspanin CD9 Determines Invasiveness and Tumorigenicity of Human Breast Cancer Cells. Oncotarget 2015, 6, 7970–7991. [Google Scholar] [CrossRef]
- Mori, M.; Mimori, K.; Shiraishi, T.; Haraguchi, M.; Ueo, H.; Barnard, G.F.; Akiyoshi, T. Motility related protein 1 (MRP1/CD9) expression in colon cancer. Clin. Cancer Res. 1998, 4, 1507–1510. [Google Scholar]
- Kim, K.J.; Kwon, H.J.; Kim, M.C.; Bae, Y.K. CD9 Expression in Colorectal Carcinomas and Its Prognostic Significance. J. Pathol. Transl. Med. 2016, 50, 459–468. [Google Scholar] [CrossRef]
- Hashida, H.; Takabayashi, A.; Tokuhara, T.; Hattori, N.; Taki, T.; Hasegawa, H.; Satoh, S.; Kobayashi, N.; Yamaoka, Y.; Miyake, M. Clinical significance of transmembrane 4 superfamily in colon cancer. Br. J. Cancer 2003, 89, 158–167. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.; Bain, L.J.; Belinsky, M.G.; Kruh, G.D. Expression of multidrug resistance protein-3 (multispecific organic anion transporter-D) in human embryonic kidney 293 cells confers resistance to anticancer agents. Cancer Res. 1999, 59, 5964–5967. [Google Scholar] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, M.F.; Rappa, G.; Fontana, S.; Karbanová, J.; Aalam, F.; Tai, D.; Li, Z.; Pucci, M.; Alessandro, R.; Morimoto, C.; et al. Anti-Human CD9 Fab Fragment Antibody Blocks the Extracellular Vesicle-Mediated Increase in Malignancy of Colon Cancer Cells. Cells 2022, 11, 2474. https://doi.org/10.3390/cells11162474
Santos MF, Rappa G, Fontana S, Karbanová J, Aalam F, Tai D, Li Z, Pucci M, Alessandro R, Morimoto C, et al. Anti-Human CD9 Fab Fragment Antibody Blocks the Extracellular Vesicle-Mediated Increase in Malignancy of Colon Cancer Cells. Cells. 2022; 11(16):2474. https://doi.org/10.3390/cells11162474
Chicago/Turabian StyleSantos, Mark F., Germana Rappa, Simona Fontana, Jana Karbanová, Feryal Aalam, Derek Tai, Zhiyin Li, Marzia Pucci, Riccardo Alessandro, Chikao Morimoto, and et al. 2022. "Anti-Human CD9 Fab Fragment Antibody Blocks the Extracellular Vesicle-Mediated Increase in Malignancy of Colon Cancer Cells" Cells 11, no. 16: 2474. https://doi.org/10.3390/cells11162474
APA StyleSantos, M. F., Rappa, G., Fontana, S., Karbanová, J., Aalam, F., Tai, D., Li, Z., Pucci, M., Alessandro, R., Morimoto, C., Corbeil, D., & Lorico, A. (2022). Anti-Human CD9 Fab Fragment Antibody Blocks the Extracellular Vesicle-Mediated Increase in Malignancy of Colon Cancer Cells. Cells, 11(16), 2474. https://doi.org/10.3390/cells11162474