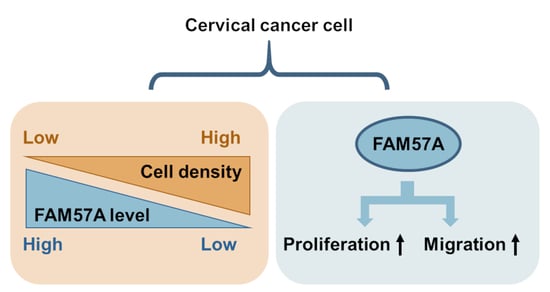
FAM57A (Family with Sequence Similarity 57 Member A) Is a Cell-Density-Regulated Protein and Promotes the Proliferation and Migration of Cervical Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. Immunoblot Analyses
2.3. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.4. Transfections with Plasmids and siRNAs
2.5. Colony Formation Assays (CFAs)
2.6. Live Cell Imaging Microscopy
2.7. Statistical Analyses
3. Results
3.1. FAM57A Protein Levels Are Highly Increased in Cells Cultivated at Hypoxia Compared to Normoxia
3.2. FAM57A Protein Levels Are Strongly Dependent on Cell Density
3.3. FAM57A Protein Levels in Hypoxic Cells Are Primarily a Function of Low Cell Density
3.4. Other Anti-Proliferative Stimuli, Such as HPV E6/E7 Oncogene Silencing, Also Result in a Relative Increase of FAM57A Protein Levels
3.5. FAM57A Promotes the Proliferation and Migration of Cervical Cancer Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Vaupel, P.; Mayer, A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007, 26, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Singleton, D.C.; Macann, A.; Wilson, W.R. Therapeutic targeting of the hypoxic tumour microenvironment. Nat. Rev. Clin. Oncol. 2021, 18, 751–772. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hoppe-Seyler, K.; Bossler, F.; Lohrey, C.; Bulkescher, J.; Rosl, F.; Jansen, L.; Mayer, A.; Vaupel, P.; Durst, M.; Hoppe-Seyler, F. Induction of dormancy in hypoxic human papillomavirus-positive cancer cells. Proc. Natl. Acad. Sci. USA 2017, 114, E990–E998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoppe-Seyler, K.; Bossler, F.; Braun, J.A.; Herrmann, A.L.; Hoppe-Seyler, F. The hpv e6/e7 oncogenes: Key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol. 2018, 26, 158–168. [Google Scholar] [CrossRef]
- Bossler, F.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Pi3k/akt/mtor signaling regulates the virus/host cell crosstalk in hpv-positive cervical cancer cells. Int. J. Mol. Sci. 2019, 20, 2188. [Google Scholar] [CrossRef] [Green Version]
- Bossler, F.; Kuhn, B.J.; Gunther, T.; Kraemer, S.J.; Khalkar, P.; Adrian, S.; Lohrey, C.; Holzer, A.; Shimobayashi, M.; Durst, M.; et al. Repression of human papillomavirus oncogene expression under hypoxia is mediated by pi3k/mtorc2/akt signaling. mBio 2019, 10, e02323-18. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Di, Y.; Li, J.; Xie, Y.; Tang, Y.; Zhang, F.; Wei, L.; Zhang, Y.; Qin, W.; Huo, K.; et al. Molecular cloning and characterization of ct120, a novel membrane-associated gene involved in amino acid transport and glutathione metabolism. Biochem. Biophys. Res. Commun. 2002, 297, 528–536. [Google Scholar] [CrossRef]
- Gene [Internet]. Bethesda (md): National library of Medicine (us), National Center for Biotechnology Information. 2004. Available online: https://www.ncbi.nlm.nih.gov/gene/79850 (accessed on 24 August 2022).
- Baltaci, E.; Ekizoglu, S.; Sari, E.; Karaman, E.; Ulutin, T.; Buyru, N. Ct120a acts as an oncogene in head and neck squamous cell carcinoma. J. Cancer 2015, 6, 1255–1259. [Google Scholar] [CrossRef] [Green Version]
- He, X.H.; Li, J.J.; Xie, Y.H.; Zhang, F.R.; Qu, S.M.; Tang, Y.T.; Qin, W.X.; Wan, D.F.; Gu, J.R. Expression of human novel gene ct120 in lung cancer and its effects on cell growth. Ai Zheng 2003, 22, 113–118. [Google Scholar]
- Li, Z.; Shao, S.; Xie, S.; Jiao, F.; Ma, Y.; Shi, S. Silencing of ct120 by antisense oligonucleotides could inhibit the lung cancer cells growth. Ir. J. Med. Sci. 2010, 179, 217–223. [Google Scholar] [CrossRef]
- Pan, D.; Wei, L.; Yao, M.; Wan, D.; Gu, J. Down-regulation of ct120a by RNA interference suppresses lung cancer cells growth and sensitizes to ultraviolet-induced apoptosis. Cancer Lett. 2006, 235, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Liu, Y.; Zhao, C. Integrated analysis of fam57a expression and its potential roles in hepatocellular carcinoma. Front. Oncol. 2021, 11, 719973. [Google Scholar] [CrossRef] [PubMed]
- Ramanand, S.G.; Chen, Y.; Yuan, J.; Daescu, K.; Lambros, M.B.; Houlahan, K.E.; Carreira, S.; Yuan, W.; Baek, G.; Sharp, A.; et al. The landscape of RNA polymerase ii-associated chromatin interactions in prostate cancer. J. Clin. Investig. 2020, 130, 3987–4005. [Google Scholar] [CrossRef]
- Emami, N.C.; Kachuri, L.; Meyers, T.J.; Das, R.; Hoffman, J.D.; Hoffmann, T.J.; Hu, D.; Shan, J.; Feng, F.Y.; Ziv, E.; et al. Association of imputed prostate cancer transcriptome with disease risk reveals novel mechanisms. Nat. Commun. 2019, 10, 3107. [Google Scholar] [CrossRef] [Green Version]
- Ren, N.; Liu, Q.; Yan, L.; Huang, Q. Parallel reporter assays identify altered regulatory role of rs684232 in leading to prostate cancer predisposition. Int. J. Mol. Sci. 2021, 22, 8792. [Google Scholar] [CrossRef] [PubMed]
- Baldus, S.E.; Schwarz, E.; Lohrey, C.; Zapatka, M.; Landsberg, S.; Hahn, S.A.; Schmidt, D.; Dienes, H.P.; Schmiegel, W.H.; Schwarte-Waldhoff, I. Smad4 deficiency in cervical carcinoma cells. Oncogene 2005, 24, 810–819. [Google Scholar] [CrossRef] [Green Version]
- Frensemeier, K.; Holzer, A.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Dickkopf-1 expression is repressed by oncogenic human papillomaviruses (hpvs) and regulates the cisplatin sensitivity of hpv-positive cancer cells in a jnk-dependent manner. Int. J. Cancer 2022, 151, 2215–2228. [Google Scholar] [CrossRef]
- Leitz, J.; Reuschenbach, M.; Lohrey, C.; Honegger, A.; Accardi, R.; Tommasino, M.; Llano, M.; von Knebel Doeberitz, M.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Oncogenic human papillomaviruses activate the tumor-associated lens epithelial-derived growth factor (ledgf) gene. PLoS Pathog. 2014, 10, e1003957. [Google Scholar] [CrossRef] [PubMed]
- Kohler, G.; Milstein, C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature 1975, 256, 495–497. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fjeldbo, C.S.; Aarnes, E.K.; Malinen, E.; Kristensen, G.B.; Lyng, H. Identification and validation of reference genes for rt-qpcr studies of hypoxia in squamous cervical cancer patients. PLoS ONE 2016, 11, e0156259. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Okayama, H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell Biol. 1987, 7, 2745–2752. [Google Scholar]
- Brummelkamp, T.R.; Bernards, R.; Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002, 296, 550–553. [Google Scholar] [CrossRef] [Green Version]
- Cullmann, C.; Hoppe-Seyler, K.; Dymalla, S.; Lohrey, C.; Scheffner, M.; Durst, M.; Hoppe-Seyler, F. Oncogenic human papillomaviruses block expression of the b-cell translocation gene-2 tumor suppressor gene. Int. J. Cancer 2009, 125, 2014–2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrmann, A.L.; Kuhn, B.J.; Holzer, A.; Krijgsveld, J.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Delineating the switch between senescence and apoptosis in cervical cancer cells under ciclopirox treatment. Cancers 2021, 13, 4995. [Google Scholar] [CrossRef]
- Vang Mouritzen, M.; Jenssen, H. Optimized scratch assay for in vitro testing of cell migration with an automated optical camera. J. Vis. Exp. 2018, 8, e57691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boukamp, P.; Petrussevska, R.T.; Breitkreutz, D.; Hornung, J.; Markham, A.; Fusenig, N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988, 106, 761–771. [Google Scholar] [CrossRef] [Green Version]
- Hubbi, M.E.; Semenza, G.L. Regulation of cell proliferation by hypoxia-inducible factors. Am. J. Physiol. Cell Physiol. 2015, 309, C775–C782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schodel, J.; Oikonomopoulos, S.; Ragoussis, J.; Pugh, C.W.; Ratcliffe, P.J.; Mole, D.R. High-resolution genome-wide mapping of hif-binding sites by chip-seq. Blood 2011, 117, e207–e217. [Google Scholar] [CrossRef] [Green Version]
- Wicks, E.E.; Semenza, G.L. Hypoxia-inducible factors: Cancer progression and clinical translation. J. Clin. Investig. 2022, 132, e159839. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.H.; Alexander, K.A. RNA interference of human papillomavirus type 18 e6 and e7 induces senescence in hela cells. J. Virol. 2003, 77, 6066–6069. [Google Scholar] [CrossRef] [PubMed]
- Hoppe-Seyler, K.; Herrmann, A.L.; Daschle, A.; Kuhn, B.J.; Strobel, T.D.; Lohrey, C.; Bulkescher, J.; Krijgsveld, J.; Hoppe-Seyler, F. Effects of metformin on the virus/host cell crosstalk in human papillomavirus-positive cancer cells. Int J. Cancer 2021, 149, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Toker, A. Akt/pkb signaling: Navigating the network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef] [Green Version]
- Lavoie, H.; Gagnon, J.; Therrien, M. Erk signalling: A master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 2020, 21, 607–632. [Google Scholar] [CrossRef]
- He, X.H.; Li, J.J.; Xie, Y.H.; Tang, Y.T.; Yao, G.F.; Qin, W.X.; Wan, D.F.; Gu, J.R. Altered gene expression profiles of nih3t3 cells regulated by human lung cancer associated gene ct120. Cell Res. 2004, 14, 487–496. [Google Scholar] [CrossRef] [Green Version]
- Baltaci, E.; Seyhan, B.; Baykara, O.; Buyru, N. Ct120: A new potential target for c-myc in head and neck cancers. J. Cancer 2017, 8, 880–886. [Google Scholar] [CrossRef] [Green Version]
- Fussbroich, B.; Wagener, N.; Macher-Goeppinger, S.; Benner, A.; Falth, M.; Sultmann, H.; Holzer, A.; Hoppe-Seyler, K.; Hoppe-Seyler, F. Ezh2 depletion blocks the proliferation of colon cancer cells. PLoS ONE 2011, 6, e21651. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.J.; Wang, L.Y.; Chodosh, L.A.; Keith, B.; Simon, M.C. Differential roles of hypoxia-inducible factor 1alpha (hif-1alpha) and hif-2alpha in hypoxic gene regulation. Mol. Cell Biol. 2003, 23, 9361–9374. [Google Scholar] [CrossRef] [Green Version]
- Copple, B.L.; Bai, S.; Burgoon, L.D.; Moon, J.O. Hypoxia-inducible factor-1alpha regulates the expression of genes in hypoxic hepatic stellate cells important for collagen deposition and angiogenesis. Liver Int. 2011, 31, 230–244. [Google Scholar] [CrossRef] [Green Version]
- Chan, M.C.; Ilott, N.E.; Schodel, J.; Sims, D.; Tumber, A.; Lippl, K.; Mole, D.R.; Pugh, C.W.; Ratcliffe, P.J.; Ponting, C.P.; et al. Tuning the transcriptional response to hypoxia by inhibiting hypoxia-inducible factor (hif) prolyl and asparaginyl hydroxylases. J. Biol. Chem. 2016, 291, 20661–20673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, M.; Triboulet, R.; Mohseni, M.; Schlegelmilch, K.; Shrestha, K.; Camargo, F.D.; Gregory, R.I. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 2014, 156, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Sharif, G.M.; Schmidt, M.O.; Yi, C.; Hu, Z.; Haddad, B.R.; Glasgow, E.; Riegel, A.T.; Wellstein, A. Cell growth density modulates cancer cell vascular invasion via hippo pathway activity and cxcr2 signaling. Oncogene 2015, 34, 5879–5889. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Meng, Z.; Chen, R.; Guan, K.L. The hippo pathway: Biology and pathophysiology. Annu. Rev. Biochem. 2019, 88, 577–604. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.C.; Wu, J.W.; Wang, C.W.; Jang, A.C. Hippo signaling-mediated mechanotransduction in cell movement and cancer metastasis. Front. Mol. Biosci. 2019, 6, 157. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wu, J.; Ling, M.T.; Zhao, L.; Zhao, K.N. The role of the pi3k/akt/mtor signalling pathway in human cancers induced by infection with human papillomaviruses. Mol. Cancer 2015, 14, 87. [Google Scholar] [CrossRef] [Green Version]
- Luna, A.J.; Sterk, R.T.; Griego-Fisher, A.M.; Chung, J.Y.; Berggren, K.L.; Bondu, V.; Barraza-Flores, P.; Cowan, A.T.; Gan, G.N.; Yilmaz, E.; et al. Mek/erk signaling is a critical regulator of high-risk human papillomavirus oncogene expression revealing therapeutic targets for HPV-induced tumors. PLoS Pathog. 2021, 17, e1009216. [Google Scholar] [CrossRef]
- Talks, K.L.; Turley, H.; Gatter, K.C.; Maxwell, P.H.; Pugh, C.W.; Ratcliffe, P.J.; Harris, A.L. The expression and distribution of the hypoxia-inducible factors hif-1alpha and hif-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am. J. Pathol. 2000, 157, 411–421. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, D.; Strobel, T.D.; Bulkescher, J.; Tessmer, C.; Hofmann, I.; Hoppe-Seyler, F.; Hoppe-Seyler, K. FAM57A (Family with Sequence Similarity 57 Member A) Is a Cell-Density-Regulated Protein and Promotes the Proliferation and Migration of Cervical Cancer Cells. Cells 2022, 11, 3309. https://doi.org/10.3390/cells11203309
Yang D, Strobel TD, Bulkescher J, Tessmer C, Hofmann I, Hoppe-Seyler F, Hoppe-Seyler K. FAM57A (Family with Sequence Similarity 57 Member A) Is a Cell-Density-Regulated Protein and Promotes the Proliferation and Migration of Cervical Cancer Cells. Cells. 2022; 11(20):3309. https://doi.org/10.3390/cells11203309
Chicago/Turabian StyleYang, Dongyun, Tobias D. Strobel, Julia Bulkescher, Claudia Tessmer, Ilse Hofmann, Felix Hoppe-Seyler, and Karin Hoppe-Seyler. 2022. "FAM57A (Family with Sequence Similarity 57 Member A) Is a Cell-Density-Regulated Protein and Promotes the Proliferation and Migration of Cervical Cancer Cells" Cells 11, no. 20: 3309. https://doi.org/10.3390/cells11203309
APA StyleYang, D., Strobel, T. D., Bulkescher, J., Tessmer, C., Hofmann, I., Hoppe-Seyler, F., & Hoppe-Seyler, K. (2022). FAM57A (Family with Sequence Similarity 57 Member A) Is a Cell-Density-Regulated Protein and Promotes the Proliferation and Migration of Cervical Cancer Cells. Cells, 11(20), 3309. https://doi.org/10.3390/cells11203309