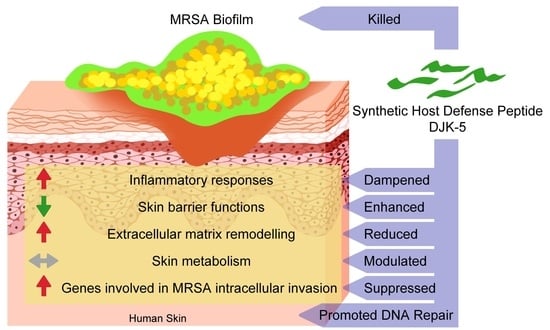
Host Response of Human Epidermis to Methicillin-Resistant Staphylococcus aureus Biofilm Infection and Synthetic Antibiofilm Peptide Treatment
Abstract
:1. Introduction
2. Material and Methods
2.1. Peptide and Reagents
2.2. Bacteria Culture
2.3. MRSA USA300-LAC Thermal Wounding ex Vivo Skin Model
2.4. H&E Staining
2.5. MRSA USA300-LAC Thermal Wounding N/TERT Skin Model
2.6. RNA Isolation
2.7. RNA-Seq Library Preparation
2.8. RNA-Seq Analysis
2.9. Statistical Analysis
3. Results
3.1. DJK-5 Reduced MRSA Biofilm and Inflammation in Thermally Wounded Human Skin
3.2. Effect of DJK-5 Treatment on MRSA Biofilm Infected Thermally-Injured Skin Transcriptome
3.3. MRSA Biofilm Infection Impaired Skin Barrier Function and Enhanced Extracellular Matrix Turnover in Thermally-Injured Skin
3.4. DJK-5 Dampened MRSA Biofilm-Induced Skin Inflammation
3.5. DJK-5 Reduced the Cellular Stress Response to Amino Acid Starvation
3.6. DJK-5 Promoted DNA Repair Function in MRSA Biofilm Infected Skin
3.7. DJK-5 Reduced Pathways Mediating MRSA Invasion of Thermally-Injured Skin
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 5 September 2022).
- WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 5 September 2022).
- De La Fuente-Núñez, C.; Cardoso, M.H.; de Souza Cândido, E.; Franco, O.L.; Hancock, R.E.W. Synthetic antibiofilm peptides. Biochim. Biophys. Acta 2016, 1858, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Ki, V.; Rotstein, C. Bacterial Skin and Soft Tissue Infections in Adults: A Review of Their Epidemiology, Pathogenesis, Diagnosis, Treatment and Site of Care. Can. J. Infect. Dis. Med. Microbiol. 2008, 19, 173–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archer, N.K.; Mazaitis, M.J.; Costerton, J.W.; Leid, J.G.; Powers, M.E.; Shirtliff, M.E. Staphylococcus aureus biofilms: Properties, regulation, and roles in human disease. Virulence 2011, 2, 445–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwiecinski, J.; Kahlmeter, G.; Jin, T. Biofilm Formation by Staphylococcus aureus Isolates from Skin and Soft Tissue Infections. Curr. Microbiol. 2015, 70, 698–703. [Google Scholar] [CrossRef]
- Yousef, H.; Alhajj, M.; Sharma, S. Anatomy, Skin (Integument), Epidermis. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: http://www.ncbi.nlm.nih.gov/books/NBK470464/ (accessed on 5 September 2022).
- Watters, C.; Fleming, D.; Bishop, D.; Rumbaugh, K. Host Responses to Biofilm. Prog. Mol. Biol. Transl. Sci. 2016, 142, 193–239. [Google Scholar] [CrossRef]
- Lowy, F.D. Is Staphylococcus aureus an intracellular pathogen? Trends Microbiol. 2000, 8, 341–343. [Google Scholar] [CrossRef]
- Mirzaei, R.; Ranjbar, R.; Karampoor, S.; Goodarzi, R.; Hasanvand, H. The Human Immune System toward Staphylococcus aureus. Open Microbiol. J. 2020, 14, 164–170. [Google Scholar] [CrossRef]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Haney, E.F.; Straus, S.K.; Hancock, R.E.W. Reassessing the Host Defense Peptide Landscape. Front. Chem. 2019, 7, 43. [Google Scholar] [CrossRef] [Green Version]
- de la Fuente-Núñez, C.; Reffuveille, F.; Mansour, S.C.; Reckseidler-Zenteno, S.L.; Hernández, D.; Brackman, G.; Coenye, T.; Hancock, R.E. D-Enantiomeric Peptides that Eradicate Wild-Type and Multidrug-Resistant Biofilms and Protect against Lethal Pseudomonas aeruginosa Infections. Chem. Biol. 2015, 22, 196–205. [Google Scholar] [CrossRef]
- Mansour, S.C. A Novel Peptide-Based Treatment for Bacterial Abscess Infections. Ph.D. Thesis, University of British Columbia, Vancouver, BC, Canada, 2018. [Google Scholar] [CrossRef]
- Pletzer, D.; Wolfmeier, H.; Bains, M.; Hancock, R.E.W. Synthetic Peptides to Target Stringent Response-Controlled Virulence in a Pseudomonas aeruginosa Murine Cutaneous Infection Model. Front. Microbiol. 2017, 8, 1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centers for Disease Control and Prevention (CDC). Outbreaks of community-associated methicillin-resistant Staphylococcus aureus skin infections-Los Angeles County, California, 2002–2003. MMWR Morb. Mortal. Wkly. Rep. 2003, 52, 88. [Google Scholar]
- de Breij, A.; Riool, M.; Cordfunke, R.A.; Malanovic, N.; de Boer, L.; Koning, R.I.; Ravensbergen, E.; Franken, M.; van der Heijde, T.; Boekema, B.K.; et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018, 10, eaan4044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, B.; Haney, E.F.; Akhoundsadegh, N.; Pletzer, D.; Trimble, M.J.; Adriaans, A.E.; Nibbering, P.H.; Hancock, R.E.W. Human organoid biofilm model for assessing antibiofilm activity of novel agents. NPJ Biofilms Microbiomes 2021, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.C.; Lee, A.; Hancock, R.E.W. Mechanisms of the Innate Defense Regulator Peptide-1002 Anti-Inflammatory Activity in a Sterile Inflammation Mouse Model. J. Immunol. 2017, 199, 3592–3603. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 September 2022).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [Green Version]
- Aken, B.L.; Ayling, S.; Barrell, D.; Clarke, L.; Curwen, V.; Fairley, S.; Banet, J.F.; Billis, K.; Girón, C.G.; Hourlier, T.; et al. The Ensembl gene annotation system. Database 2016, 2016, baw093. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; He, Q.-Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. BioSyst. 2016, 12, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Foroushani, A.B.; Brinkman, F.; Lynn, D.J. Pathway-GPS and SIGORA: Identifying relevant pathways based on the over-representation of their gene-pair signatures. PeerJ 2013, 1, e229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, J.; Gill, E.E.; Hancock, R.E.W. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat. Protoc. 2015, 10, 823–844. [Google Scholar] [CrossRef] [PubMed]
- Piipponen, M.; Li, D.; Landén, N.X. The Immune Functions of Keratinocytes in Skin Wound Healing. Int. J. Mol. Sci. 2020, 21, 8790. [Google Scholar] [CrossRef]
- Elias, P.M. The skin barrier as an innate immune element. Semin. Immunopathol. 2007, 29, 3–14. [Google Scholar] [CrossRef]
- Bragulla, H.H.; Homberger, D.G. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J. Anat. 2009, 214, 516–559. [Google Scholar] [CrossRef]
- Tomlin, H.; Piccinini, A.M. A complex interplay between the extracellular matrix and the innate immune response to microbial pathogens. Immunology 2018, 155, 186–201. [Google Scholar] [CrossRef] [Green Version]
- Pfisterer, K.; Shaw, L.E.; Symmank, D.; Weninger, W. The Extracellular Matrix in Skin Inflammation and Infection. Front. Cell Dev. Biol. 2021, 9, 682414. [Google Scholar] [CrossRef]
- Rodrigues, L.O.C.P.; Graça, R.S.F.; Carneiro, L.A.M. Integrated Stress Responses to Bacterial Pathogenesis Patterns. Front. Immunol. 2018, 9, 1306. [Google Scholar] [CrossRef]
- Wek, R.C. Role of eIF2α Kinases in Translational Control and Adaptation to Cellular Stress. Cold Spring Harb. Perspect. Biol. 2018, 10, a032870. [Google Scholar] [CrossRef] [PubMed]
- Nakad, R.; Schumacher, B. DNA Damage Response and Immune Defense: Links and Mechanisms. Front. Genet. 2016, 7, 147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babushkina, N.P.; Postrigan, A.E.; Kucher, A.N. Involvement of Variants in the Genes Encoding BRCA1-Associated Genome Surveillance Complex (BASC) in the Development of Human Common Diseases. Mol. Biol. 2021, 55, 318–337. [Google Scholar] [CrossRef]
- Ranjha, L.; Howard, S.M.; Cejka, P. Main steps in DNA double-strand break repair: An introduction to homologous recombination and related processes. Chromosoma 2018, 127, 187–214. [Google Scholar] [CrossRef] [Green Version]
- Marbach, H.; Vizcay-Barrena, G.; Memarzadeh, K.; Otter, J.A.; Pathak, S.; Allaker, R.P.; Harvey, R.D.; Edgeworth, J.D. Tolerance of MRSA ST239-TW to chlorhexidine-based decolonization: Evidence for keratinocyte invasion as a mechanism of biocide evasion. J. Infect. 2019, 78, 119–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rollin, G.; Tan, X.; Tros, F.; Dupuis, M.; Nassif, X.; Charbit, A.; Coureuil, M. Intracellular Survival of Staphylococcus aureus in Endothelial Cells: A Matter of Growth or Persistence. Front. Microbiol. 2017, 8, 1354. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Tsoi, L.C.; Billi, A.C.; Ward, N.L.; Harms, P.W.; Zeng, C.; Maverakis, E.; Kahlenberg, J.M.; Gudjonsson, J.E. Cytokinocytes: The diverse contribution of keratinocytes to immune responses in skin. JCI Insight 2020, 5, e142067. [Google Scholar] [CrossRef]
- Metcalf, D.G.; Bowler, P.G. Biofilm delays wound healing: A review of the evidence. Burn. Trauma 2015, 1, 5–12. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.; Usui, M.L.; Lippman, S.I.; James, G.A.; Stewart, P.S.; Fleckman, P.; Olerud, J.E. Biofilms and Inflammation in Chronic Wounds. Adv. Wound Care 2013, 2, 389–399. [Google Scholar] [CrossRef] [Green Version]
- Mottola, C.; Matias, C.S.; Mendes, J.J.; Melo-Cristino, J.; Tavares, L.; Cavaco-Silva, P.; Oliveira, M. Susceptibility patterns of Staphylococcus aureus biofilms in diabetic foot infections. BMC Microbiol. 2016, 16, 1–9. [Google Scholar] [CrossRef]
- Du, W.; Lenz, D.; Köhler, R.; Zhang, E.; Cendon, C.; Li, J.; Massoud, M.; Wachtlin, J.; Bodo, J.; Hauser, A.E.; et al. Rapid Isolation of Functional ex vivo Human Skin Tissue-Resident Memory T Lymphocytes. Front. Immunol. 2021, 12, 624013. [Google Scholar] [CrossRef] [PubMed]
- He, X.; de Oliveira, V.L.; Keijsers, R.; Joosten, I.; Koenen, H.J. Lymphocyte Isolation from Human Skin for Phenotypic Analysis and Ex Vivo Cell Culture. J. Vis. Exp. 2016, 110, e52564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- den Reijer, P.M.; Haisma, E.M.; Lemmens-den Toom, N.A.; Willemse, J.; Koning, R.I.; Demmers, J.A.; Dekkers, D.H.; Rijkers, E.; El Ghalbzouri, A.; Nibbering, P.H.; et al. Detection of Alpha-Toxin and Other Virulence Factors in Biofilms of Staphylococcus aureus on Polystyrene and a Human Epidermal Model. PLoS ONE 2016, 11, e0145722. [Google Scholar] [CrossRef]
- Allen, H.B.; Vaze, N.D.; Choi, C.; Hailu, T.; Tulbert, B.H.; Cusack, C.A.; Joshi, S.G. The Presence and Impact of Biofilm-Producing Staphylococci in Atopic Dermatitis. JAMA Dermatol. 2014, 150, 260–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez, T.; Myers, J.M.B.; Herr, A.B.; Hershey, G.K.K. Staphylococcal Biofilms in Atopic Dermatitis. Curr. Allergy Asthma Rep. 2017, 17, 81. [Google Scholar] [CrossRef]
- Kuo, C.; Lim, S.; King, N.J.C.; Johnston, S.L.; Burgess, J.; Black, J.L.; Oliver, B.G. Rhinovirus infection induces extracellular matrix protein deposition in asthmatic and nonasthmatic airway smooth muscle cells. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2011, 300, L951–L957. [Google Scholar] [CrossRef] [PubMed]
- Mezentsev, A.; Nikolaev, A.; Bruskin, S. Matrix metalloproteinases and their role in psoriasis. Gene 2014, 540, 1–10. [Google Scholar] [CrossRef]
- Parnham, A.; Bousfield, C. The influence of matrix metalloproteases and biofilm on chronic wound healing: A discussion. Br. J. Community Nurs. 2018, 23, S22–S29. [Google Scholar] [CrossRef]
- Cho, J.S.; Pietras, E.M.; Garcia, N.C.; Ramos, R.I.; Farzam, D.M.; Monroe, H.R.; Magorien, J.E.; Blauvelt, A.; Kolls, J.K.; Cheung, A.L.; et al. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J. Clin. Investig. 2010, 120, 1762–1773. [Google Scholar] [CrossRef] [Green Version]
- Thammavongsa, V.; Kim, H.K.; Missiakas, D.; Schneewind, O. Staphylococcal manipulation of host immune responses. Nature Rev. Microbiol. 2015, 13, 529–543. [Google Scholar] [CrossRef]
- Pätzold, L.; Stark, A.; Ritzmann, F.; Meier, C.; Tschernig, T.; Reichrath, J.; Bals, R.; Bischoff, M.; Beisswenger, C. IL-17C and IL-17RE Promote Wound Closure in a Staphylococcus aureus-Based Murine Wound Infection Model. Microorganisms 2021, 9, 1821. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, S.; Ying, S.; Tang, S.; Ding, Y.; Li, Y.; Qiao, J.; Fang, H. The IL-23/IL-17 Pathway in Inflammatory Skin Diseases: From Bench to Bedside. Front. Immunol. 2020, 11, 594735. [Google Scholar] [CrossRef] [PubMed]
- Piskin, G.; Sylva-Steenland, R.M.R.; Bos, J.D.; Teunissen, M.B.M. In Vitro and In Situ Expression of IL-23 by Keratinocytes in Healthy Skin and Psoriasis Lesions: Enhanced Expression in Psoriatic Skin. J. Immunol. 2006, 176, 1908–1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauffer, F.; Jargosch, M.; Baghin, V.; Krause, L.; Kempf, W.; Absmaier-Kijak, M.; Morelli, M.; Madonna, S.; Marsais, F.; Lepescheux, L.; et al. IL-17C amplifies epithelial inflammation in human psoriasis and atopic eczema. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Waring, P.M.; Waring, L.J.; Billington, T.; Metcalf, D. Leukemia inhibitory factor protects against experimental lethal Escherichia coli septic shock in mice. Proc. Natl. Acad. Sci. USA 1995, 92, 1337–1341. [Google Scholar] [CrossRef] [Green Version]
- West, N.R. Coordination of Immune-Stroma Crosstalk by IL-6 Family Cytokines. Front. Immunol. 2019, 10, 1093. [Google Scholar] [CrossRef]
- Moser, C.; Pedersen, H.T.; Lerche, C.J.; Kolpen, M.; Line, L.; Thomsen, K.; Høiby, N.; Jensen, P. Biofilms and host response—Helpful or harmful. APMIS 2017, 125, 320–338. [Google Scholar] [CrossRef] [Green Version]
- Pena, O.M.; Afacan, N.; Pistolic, J.; Chen, C.; Madera, L.; Falsafi, R.; Fjell, C.; Hancock, R. Synthetic Cationic Peptide IDR-1018 Modulates Human Macrophage Differentiation. PLoS ONE 2013, 8, e52449. [Google Scholar] [CrossRef] [Green Version]
- Deplanche, M.; Mouhali, N.; Nguyen, M.-T.; Cauty, C.; Ezan, F.; Diot, A.; Raulin, L.; Dutertre, S.; Langouet, S.; Legembre, P.; et al. Staphylococcus aureus induces DNA damage in host cell. Sci. Rep. 2019, 9, 7694. [Google Scholar] [CrossRef] [Green Version]
- Krueger, A.; Mohamed, A.; Kolka, C.M.; Stoll, T.; Zaugg, J.; Linedale, R.; Morrison, M.; Soyer, H.P.; Hugenholtz, P.; Frazer, I.H.; et al. Skin Cancer-Associated S. aureus Strains Can Induce DNA Damage in Human Keratinocytes by Downregulating DNA Repair and Promoting Oxidative Stress. Cancers 2022, 14, 2143. [Google Scholar] [CrossRef]
- Maréchal, A.; Zou, L. DNA Damage Sensing by the ATM and ATR Kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef] [PubMed]
- Dray, E.; Etchin, J.; Wiese, C.; Saro, D.; Williams, G.J.; Hammel, M.; Yu, X.; E Galkin, V.; Liu, D.; Tsai, M.-S.; et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 2010, 17, 1255–1259. [Google Scholar] [CrossRef] [PubMed]
- Sy, S.M.H.; Huen, M.S.Y.; Chen, J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. USA 2009, 106, 7155–7160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, R.; Lavrik, O.I.; Kim, S.-J.; Kedar, P.; Yang, X.-P.; Berg, B.J.V.; Wilson, S.H. DNA Polymerase β-mediated Long Patch Base Excision Repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem. 2001, 276, 32411–32414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Josse, J.; Laurent, F.; Diot, A. Staphylococcal Adhesion and Host Cell Invasion: Fibronectin-Binding and Other Mechanisms. Front. Microbiol. 2017, 8, 2433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larjava, H.; Koivisto, L.; Häkkinen, L. Keratinocyte Interactions with Fibronectin during Wound Healing. Madame Curie Bioscience Database [Internet]. Landes Bioscience. 2013. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6391/ (accessed on 5 September 2022).
- Fowler, T.; Johansson, S.; Wary, K.K.; Hook, M. Src kinase has a central role in in vitro cellular internalization of Staphylococcus aureus. Cell. Microbiol. 2003, 5, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Agerer, F.; Lux, S.; Michel, A.; Rohde, M.; Ohlsen, K.; Hauck, C.R. Cellular invasion by Staphylococcus aureus reveals a functional link between focal adhesion kinase and cortactin in integrin-mediated internalisation. J. Cell Sci. 2005, 118, 2189–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.-H.; Zhang, K.; Wang, N.; Qiu, X.-M.; Wang, Y.-B.; He, P. Involvement of phosphatidylinositol 3-Kinase/Akt signaling pathway in β1 integrin-mediated internalization of Staphylococcus aureus by alveolar epithelial cells. J. Microbiol. 2013, 51, 644–650. [Google Scholar] [CrossRef]
- Shen, Y.; Naujokas, M.; Park, M.; Ireton, K. InlB-Dependent Internalization of Listeria Is Mediated by the Met Receptor Tyrosine Kinase. Cell 2000, 103, 501–510. [Google Scholar] [CrossRef]





| Function | Pathway | Direction | Adjusted p-Value |
|---|---|---|---|
| MRSA biofilm infected burned skin vs. burned skin control | |||
| Keratinization | Formation of the cornified envelope | down | 1.1 × 10−2 |
| Cell junction organization | Cell-extracellular matrix interactions | up | 2.2 × 10−4 |
| Extracellular matrix organization | Anchoring fibril formation | up | 7.2 × 10−3 |
| Crosslinking of collagen fibrils | up | 3.5 × 10−2 | |
| Degradation of the extracellular matrix | up | 8.6 ×10−4 | |
| Collagen degradation | up | 1.0 × 10−2 | |
| Infectious disease | Cell recruitment (pro-inflammatory response) | up | 3.5 × 10−2 |
| Innate immune system | Toll-like receptor cascades | up | 1.9 × 10−3 |
| MAP kinase activation | up | 1.9 × 10−3 | |
| Cytokine signalling in immune system | Interleukin-17 signalling | up | 3.2 × 10−3 |
| Interleukin-12 family signalling | up | 3.5 × 10−2 | |
| Interleukin-6 family signalling | up | 7.7 × 10−3 | |
| Signal transduction | GPCR downstream signalling | up | 6.0 × 10−8 |
| Death receptor signalling | up | 3.3 × 10−3 | |
| RHO GTPase cycle | up | 7.7 × 10−3 | |
| Metabolism of RNA | rRNA processing | up | 4.3 × 10−9 |
| Metabolism of proteins | Translation | up | 2.1 × 10−4 |
| Post-translational protein phosphorylation | up | 5.1 × 10−3 | |
| Metabolism of amino acids | Metabolism of amino acids and derivatives | up | 1.1 × 10−2 |
| Metabolism of lipids | Peroxisomal lipid metabolism | down | 1.1 × 10−2 |
| DJK-5 treated MRSA biofilm infected burned skin vs. biofilm infected burned skin | |||
| Keratinization | Formation of the cornified envelope | up | 1.3 × 10−2 |
| Cell junction organization | Cell-extracellular matrix interactions | down | 2.6 × 10-2 |
| Extracellular matrix organization | Anchoring fibril formation | down | 5.3 × 10−3 |
| Crosslinking of collagen fibrils | down | 3.0 × 10−2 | |
| Collagen degradation | down | 3.7 × 10−2 | |
| Toll-like receptor cascades | TLR3 cascade | down | 4.6 × 10−2 |
| TLR9 cascade | down | 3.8 × 10−2 | |
| Cytokine signalling in immune system | IL-12 family signalling | down | 8.6 × 10−3 |
| IL-6 family signalling | down | 5.3 × 10−3 | |
| Signal transduction | Signalling by MET | down | 8.4 × 10−4 |
| Signalling by ERBB2 | down | 5.1 × 10−3 | |
| Death receptor signalling | down | 1.2 × 10−2 | |
| GPCR downstream signalling | down | 1.3 × 10−5 | |
| Cell cycle | Activation of ATR in response to replication stress | up | 4.6 × 10−2 |
| DNA repair | Homologous DNA pairing and strand exchange | up | 2.9 × 10−2 |
| Resolution of abasic sites | up | 4.6 × 10−2 | |
| Metabolism | rRNA processing | down | 9.5 × 10−13 |
| Translation | down | 2.8 × 10−11 | |
| Post-translational protein phosphorylation | down | 4.7 × 10−3 | |
| Metabolism of amino acids and derivatives | down | 2.0 × 10−4 | |
| Peroxisomal lipid metabolism | up | 1.7 × 10−2 | |
| Cellular responses to stress | Cellular response to starvation | down | 5.4 × 10v21 |
| Response of EIF2AK4 to amino acid deficiency | down | 1.2 × 10−36 | |
| DJK-5 treated MRSA biofilm infected burned skin vs. burned skin control | |||
| Cytokine signalling in immune system | IL-10 signalling | up | 5.2 × 10−6 |
| IL-4 and IL-13 signalling | up | 2.2 × 10−3 | |
| Signalling by GPCR | Chemokine receptors bind chemokines | up | 4.2 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, B.; Blimkie, T.M.; Haney, E.F.; Falsafi, R.; Akhoundsadegh, N.; Hancock, R.E.W. Host Response of Human Epidermis to Methicillin-Resistant Staphylococcus aureus Biofilm Infection and Synthetic Antibiofilm Peptide Treatment. Cells 2022, 11, 3459. https://doi.org/10.3390/cells11213459
Wu B, Blimkie TM, Haney EF, Falsafi R, Akhoundsadegh N, Hancock REW. Host Response of Human Epidermis to Methicillin-Resistant Staphylococcus aureus Biofilm Infection and Synthetic Antibiofilm Peptide Treatment. Cells. 2022; 11(21):3459. https://doi.org/10.3390/cells11213459
Chicago/Turabian StyleWu, Bing (Catherine), Travis M. Blimkie, Evan F. Haney, Reza Falsafi, Noushin Akhoundsadegh, and Robert E. W. Hancock. 2022. "Host Response of Human Epidermis to Methicillin-Resistant Staphylococcus aureus Biofilm Infection and Synthetic Antibiofilm Peptide Treatment" Cells 11, no. 21: 3459. https://doi.org/10.3390/cells11213459
APA StyleWu, B., Blimkie, T. M., Haney, E. F., Falsafi, R., Akhoundsadegh, N., & Hancock, R. E. W. (2022). Host Response of Human Epidermis to Methicillin-Resistant Staphylococcus aureus Biofilm Infection and Synthetic Antibiofilm Peptide Treatment. Cells, 11(21), 3459. https://doi.org/10.3390/cells11213459