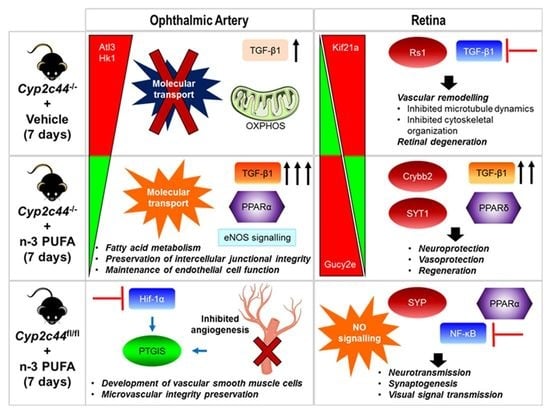
Short-Term Omega-3 Supplementation Modulates Novel Neurovascular and Fatty Acid Metabolic Proteome Changes in the Retina and Ophthalmic Artery of Mice with Targeted Cyp2c44 Gene Deletion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Experimental Design
2.3. Sample Preparation
2.4. Tissue Protein Extraction
2.5. Nano-Liquid Chromatography–Electrospray Ionization–MS/MS (nLC-ESI-MS/MS) Analysis
2.6. Label-Free Quantification (LFQ) Analysis
2.7. Statistical and Bioinformatics Analysis
3. Results
3.1. General Observation and ω-3 PUFA-Related Body Weight Changes
3.2. Label-Free Quantitative Proteomics Analysis
3.3. Influence of ω-3 PUFA Administration on the Ophthalmic Arterial Proteome
3.4. Effects of ω-3 PUFA Supplementation on the Retinal Proteome
3.5. The Role of Predicted Upstream Regulators in the Ophthalmic Artery
3.6. The Role of Predicted Upstream Regulators in the Retina
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gong, Y.; Fu, Z.; Liegl, R.; Chen, J.; Hellström, A.; Smith, L.E. ω-3 and ω-6 long-chain PUFAs and their enzymatic metabolites in neovascular eye diseases. Am. J. Clin. Nutr. 2017, 106, 16–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prokopiou, E.; Kolovos, P.; Georgiou, C.; Kalogerou, M.; Potamiti, L.; Sokratous, K.; Kyriacou, K.; Georgiou, T. Omega-3 fatty acids supplementation protects the retina from age-associated degeneration in aged C57BL/6J mice. BMJ Open Ophthalmol. 2019, 4, e000326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prokopiou, E.; Kolovos, P.; Kalogerou, M.; Neokleous, A.; Papagregoriou, G.; Deltas, C.; Malas, S.; Georgiou, T. Therapeutic potential of omega-3 fatty acids supplementation in a mouse model of dry macular degeneration. BMJ Open Ophthalmol. 2017, 1, e000056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tourtas, T.; Birke, M.T.; Kruse, F.E.; Welge-Lüssen, U.-C.; Birke, K. Preventive effects of omega-3 and omega-6 Fatty acids on peroxide mediated oxidative stress responses in primary human trabecular meshwork cells. PLoS ONE 2012, 7, e31340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arterburn, L.M.; Hall, E.B.; Oken, H. Distribution, interconversion, and dose response of n − 3 fatty acids in humans. Am. J. Clin. Nutr. 2006, 83, 1467S–1476S. [Google Scholar] [CrossRef] [Green Version]
- Bush, R.A.; Malnoe, A.; Reme, C.E.; Williams, T.P. Dietary deficiency of N-3 fatty acids alters rhodopsin content and function in the rat retina. Investig. Ophthalmol. Vis. Sci. 1994, 35, 91–100. [Google Scholar]
- Stinson, A.M.; Wiegand, R.; Anderson, R. Recycling of docosahexaenoic acid in rat retinas during n-3 fatty acid deficiency. J. Lipid Res. 1991, 32, 2009–2017. [Google Scholar] [CrossRef]
- Downie, L.E.; Vingrys, A.J. Oral omega-3 supplementation lowers intraocular pressure in normotensive adults. Transl. Vis. Sci. Technol. 2018, 7, 1. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, C.T.; Bui, B.V.; Sinclair, A.J.; Vingrys, A.J. Dietary omega 3 fatty acids decrease intraocular pressure with age by increasing aqueous outflow. Investig. Ophthalmol. Vis. Sci. 2007, 48, 756–762. [Google Scholar] [CrossRef]
- Schnebelen, C.; Pasquis, B.; Salinas-Navarro, M.; Joffre, C.; Creuzot-Garcher, C.P.; Vidal-Sanz, M.; Bron, A.M.; Bretillon, L.; Acar, N. A dietary combination of omega-3 and omega-6 polyunsaturated fatty acids is more efficient than single supplementations in the prevention of retinal damage induced by elevation of intraocular pressure in rats. Graefe’s Arch. Clin. Exp. Ophthalmol. 2009, 247, 1191–1203. [Google Scholar] [CrossRef]
- Huang, W.-B.; Fan, Q.; Zhang, X.-L. Cod liver oil: A potential protective supplement for human glaucoma. Int. J. Ophthalmol. 2011, 4, 648–651. [Google Scholar] [PubMed]
- Christen, W.G.; Schaumberg, D.A.; Glynn, R.J.; Buring, J.E. Dietary ω-3 fatty acid and fish intake and incident age-related macular degeneration in women. Arch. Ophthalmol. 2011, 129, 921–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawczynski, J.; Jentsch, S.; Schweitzer, D.; Hammer, M.; Lang, G.E.; Strobel, J. Long term effects of lutein, zeaxanthin and omega-3-LCPUFAs supplementation on optical density of macular pigment in AMD patients: The LUTEGA study. Graefe’s Arch. Clin. Exp. Ophthalmol. 2013, 251, 2711–2723. [Google Scholar] [CrossRef] [PubMed]
- García-Layana, A.; Recalde, S.; Alamán, A.S.; Robredo, P.F. Effects of lutein and docosahexaenoic acid supplementation on macular pigment optical density in a randomized controlled trial. Nutrients 2013, 5, 543–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murayama, K.; Yoneya, S.; Miyauchi, O.; Adachi-Usami, E.; Nishikawa, M. Fish oil (polyunsaturated fatty acid) prevents ischemic-induced injury in the mammalian retina. Exp. Eye Res. 2002, 74, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, T.; Wen, Y.-T.; Chang, C.-H.; Kolovos, P.; Kalogerou, M.; Prokopiou, E.; Neokleous, A.; Huang, C.-T.; Tsai, R.-K. Neuroprotective effects of omega-3 polyunsaturated fatty acids in a rat model of anterior ischemic optic neuropathy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 1603–1611. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.S.; Wang, J.J.; Flood, V.; Mitchell, P. Dietary fatty acids and the 10-year incidence of age-related macular degeneration: The Blue Mountains Eye Study. Arch. Ophthalmol. 2009, 127, 656–665. [Google Scholar] [CrossRef] [Green Version]
- Ho, L.; van Leeuwen, R.; Witteman, J.C.; van Duijn, C.M.; Uitterlinden, A.G.; Hofman, A.; de Jong, P.T.; Vingerling, J.R.; Klaver, C.C. Reducing the genetic risk of age-related macular degeneration with dietary antioxidants, zinc, and ω-3 fatty acids: The Rotterdam study. Arch. Ophthalmol. 2011, 129, 758–766. [Google Scholar] [CrossRef] [Green Version]
- Querques, G.; Benlian, P.; Chanu, B.; Portal, C.; Coscas, G.; Soubrane, G.; Souied, E. Nutritional AMD treatment phase I (NAT-1): Feasibility of oral DHA supplementation in age-related macular degeneration. Eur. J. Ophthalmol. 2009, 19, 100–106. [Google Scholar] [CrossRef]
- Sala-Vila, A.; Díaz-López, A.; Valls-Pedret, C.; Cofán, M.; García-Layana, A.; Lamuela-Raventós, R.-M.; Castañer, O.; Zanon-Moreno, V.; Martinez-Gonzalez, M.A.; Toledo, E. Dietary marine ω-3 fatty acids and incident sight-threatening retinopathy in middle-aged and older individuals with type 2 diabetes: Prospective investigation from the PREDIMED trial. JAMA Ophthalmol. 2016, 134, 1142–1149. [Google Scholar] [CrossRef] [Green Version]
- Arnold, C.; Markovic, M.; Blossey, K.; Wallukat, G.; Fischer, R.; Dechend, R.; onkel, A.; von Schacky, C.; Luft, F.C.; Muller, D.N. Arachidonic acid-metabolizing cytochrome P450 enzymes are targets of ω-3 fatty acids. J. Biol. Chem. 2010, 285, 32720–32733. [Google Scholar] [CrossRef] [PubMed]
- Méndez, L.; Ciordia, S.; Fernández, M.S.; Juárez, S.; Ramos, A.; Pazos, M.; Gallardo, J.M.; Torres, J.L.; Nogués, M.R.; Medina, I. Changes in liver proteins of rats fed standard and high-fat and sucrose diets induced by fish omega-3 PUFAs and their combination with grape polyphenols according to quantitative proteomics. J. Nutr. Biochem. 2017, 41, 84–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanai, R.; Mulki, L.; Hasegawa, E.; Takeuchi, K.; Sweigard, H.; Suzuki, J.; Gaissert, P.; Vavvas, D.G.; Sonoda, K.-H.; Rothe, M. Cytochrome P450-generated metabolites derived from ω-3 fatty acids attenuate neovascularization. Proc. Natl. Acad. Sci. USA 2014, 111, 9603–9608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amirmokhtari, N.; Foresi, B.D.; Dewan, S.S.; Bouhenni, R.A.; Smith, M.A. Absence of cytochrome P450-1b1 increases susceptibility of pressure-induced axonopathy in the murine retinal projection. Front. Cell Dev. Biol. 2021, 9, 636321. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, M.E.; McCollum, G.W.; Penn, J.S. The role of cytochrome P450 epoxygenases in retinal angiogenesis. Investig. Ophthalmol. Vis. Sci. 2014, 55, 4253–4260. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Geyer, A.; Dziumbla, S.; Awwad, K.; Zeldin, D.C.; Schunck, W.-H.; Popp, R.; Frömel, T.; Fleming, I. Role of Müller cell cytochrome P450 2c44 in murine retinal angiogenesis. Prostaglandins Other Lipid Mediat. 2017, 133, 93–102. [Google Scholar] [CrossRef]
- Shao, Z.; Fu, Z.; Stahl, A.; Joyal, J.-S.; Hatton, C.; Juan, A.; Hurst, C.; Evans, L.; Cui, Z.; Pei, D. Cytochrome P450 2C8 ω3-long-chain polyunsaturated fatty acid metabolites increase mouse retinal pathologic neovascularization—Brief report. Arterioscler.Thromb. Vasc. Biol. 2014, 34, 581–586. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Sisignano, M.; Brecht, R.; Perumal, N.; Angioni, C.; Bibli, I.-S.; Fisslthaler, B.; Kleinert, H.; Pfeiffer, N.; Fleming, I. Cyp2c44 epoxygenase-derived epoxyeicosatrienoic acids in vascular smooth muscle cells elicit vasoconstriction of the murine ophthalmic artery. Sci. Rep. 2021, 11, 18764. [Google Scholar] [CrossRef]
- Manicam, C.; Staubitz, J.; Brochhausen, C.; Grus, F.H.; Pfeiffer, N.; Gericke, A. The gatekeepers in the mouse ophthalmic artery: Endothelium-dependent mechanisms of cholinergic vasodilation. Sci. Rep. 2016, 6, 20322. [Google Scholar] [CrossRef] [Green Version]
- Perumal, N.; Straßburger, L.; Herzog, D.P.; Müller, M.B.; Pfeiffer, N.; Grus, F.H.; Manicam, C. Bioenergetic shift and actin cytoskeleton remodelling as acute vascular adaptive mechanisms to angiotensin II in murine retina and ophthalmic artery. Redox Biol. 2020, 34, 101597. [Google Scholar] [CrossRef]
- Perumal, N.; Straßburger, L.; Schmelter, C.; Gericke, A.; Pfeiffer, N.; Grus, F.H.; Manicam, C. Sample preparation for mass-spectrometry-based proteomics analysis of ocular microvessels. JoVE (J. Vis. Exp.) 2019, e59140. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Alhaider, A.A.; Bayoumy, N.; Argo, E.; Gader, A.G.; Stead, D.A. Survey of the camel urinary proteome by shotgun proteomics using a multiple database search strategy. Proteomics 2012, 12, 3403–3406. [Google Scholar] [CrossRef]
- Alpi, E.; Griss, J.; da Silva, A.W.S.; Bely, B.; Antunes, R.; Zellner, H.; Ríos, D.; O’Donovan, C.; Vizcaíno, J.A.; Martin, M.J. Analysis of the tryptic search space in UniProt databases. Proteomics 2015, 15, 48–57. [Google Scholar] [CrossRef] [Green Version]
- Shanmugam, A.K.; Nesvizhskii, A.I. Effective leveraging of targeted search spaces for improving peptide identification in tandem mass spectrometry based proteomics. J. Proteome Res. 2015, 14, 5169–5178. [Google Scholar] [CrossRef] [Green Version]
- Tanca, A.; Palomba, A.; Fraumene, C.; Pagnozzi, D.; Manghina, V.; Deligios, M.; Muth, T.; Rapp, E.; Martens, L.; Addis, M.F. The impact of sequence database choice on metaproteomic results in gut microbiota studies. Microbiome 2016, 4, 51. [Google Scholar] [CrossRef] [Green Version]
- Prokopiou, E.; Kolovos, P.; Kalogerou, M.; Neokleous, A.; Nicolaou, O.; Sokratous, K.; Kyriacou, K.; Georgiou, T. Omega-3 fatty acids supplementation: Therapeutic potential in a mouse model of Stargardt disease. Investig. Ophthalmol. Vis. Sci. 2018, 59, 2757–2767. [Google Scholar] [CrossRef] [Green Version]
- Pan, M.; Zhao, F.; Xie, B.; Wu, H.; Zhang, S.; Ye, C.; Guan, Z.; Kang, L.; Zhang, Y.; Zhou, X. Dietary ω-3 polyunsaturated fatty acids are protective for myopia. Proc. Natl. Acad. Sci. USA 2021, 118, e2104689118. [Google Scholar] [CrossRef] [PubMed]
- Ahmad Sopian, N.F.; Ajat, M.; Shafie, N.I.; Mohd Noor, M.H.; Ebrahimi, M.; Rajion, M.A.; Meng, G.Y.; Ahmad, H. Does short-term dietary omega-3 fatty acid supplementation influence brain hippocampus gene expression of zinc transporter-3? Int. J. Mol. Sci. 2015, 16, 15800–15810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ginty, A.T.; Conklin, S.M. Short-term supplementation of acute long-chain omega-3 polyunsaturated fatty acids may alter depression status and decrease symptomology among young adults with depression: A preliminary randomized and placebo controlled trial. Psychiatry Res. 2015, 229, 485–489. [Google Scholar] [CrossRef]
- Grenon, S.M.; Owens, C.D.; Nosova, E.V.; Hughes-Fulford, M.; Alley, H.F.; Chong, K.; Perez, S.; Yen, P.K.; Boscardin, J.; Hellmann, J. Short-Term, High-Dose Fish Oil Supplementation Increases the Production of Omega-3 Fatty Acid–Derived Mediators in Patients with Peripheral Artery Disease (the OMEGA-PAD I Trial). J. Am. Heart Assoc. 2015, 4, e002034. [Google Scholar] [CrossRef] [Green Version]
- Bhargava, R.; Kumar, P.; Arora, Y. Short-term omega 3 fatty acids treatment for dry eye in young and middle-aged visual display terminal users. Eye Contact Lens Sci. Clin. Pract. 2016, 42, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chen, H.; Anderson, R.E. Biosynthesis of docosahexaenoate-containing glycerolipid molecular species in the retina. J. Mol. Neurosci. 2001, 16, 205–214. [Google Scholar] [CrossRef]
- Allam-Ndoul, B.; Guénard, F.; Barbier, O.; Vohl, M.-C. Effect of different concentrations of omega-3 fatty acids on stimulated THP-1 macrophages. Genes Nutr. 2017, 12, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradberry, J.C.; Hilleman, D.E. Overview of omega-3 fatty acid therapies. Pharm. Ther. 2013, 38, 681. [Google Scholar]
- Behrendt, L.; Kurth, I.; Kaether, C. A disease causing ATLASTIN 3 mutation affects multiple endoplasmic reticulum-related pathways. Cell. Mol. Life Sci. 2019, 76, 1433–1445. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.E.; Yuan, H.; Liang, F.-X.; Sehgal, P.B. Nitric oxide scavenging causes remodeling of the endoplasmic reticulum, Golgi apparatus and mitochondria in pulmonary arterial endothelial cells. Nitric Oxide 2013, 33, 64–73. [Google Scholar] [CrossRef] [Green Version]
- Lü, L.; Niu, L.; Hu, J. “At last in” the physiological roles of the tubular ER network. Biophys. Rep. 2020, 6, 105–114. [Google Scholar] [CrossRef]
- Cao, Z.; Hao, Y.; Fung, C.W.; Lee, Y.Y.; Wang, P.; Li, X.; Xie, K.; Lam, W.J.; Qiu, Y.; Tang, B.Z. Dietary fatty acids promote lipid droplet diversity through seipin enrichment in an ER subdomain. Nat. Commun. 2019, 10, 2902. [Google Scholar] [CrossRef] [PubMed]
- López, D.; Orta, X.; Casós, K.; Sáiz, M.P.; Puig-Parellada, P.; Farriol, M.; Mitjavila, M.T. Upregulation of endothelial nitric oxide synthase in rat aorta after ingestion of fish oil-rich diet. Am. J. Physiol. -Heart Circ. Physiol. 2004, 287, H567–H572. [Google Scholar] [CrossRef] [PubMed]
- Leung, S.; Sum, W.; Shi, Y. The glycolytic process in endothelial cells and its implications. Acta Pharmacol. Sin. 2022, 43, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Shen, W.; Du, J.; Gillies, M.C. Selective knockdown of hexokinase 2 in rods leads to age-related photoreceptor degeneration and retinal metabolic remodeling. Cell Death Dis. 2020, 11, 885. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.S.; Koboldt, D.C.; Bowne, S.J.; Lang, S.; Blanton, S.H.; Cadena, E.; Avery, C.E.; Lewis, R.A.; Webb-Jones, K.; Wheaton, D.H. A dominant mutation in hexokinase 1 (HK1) causes retinitis pigmentosa. Investig. Ophthalmol. Vis. Sci. 2014, 55, 7147–7158. [Google Scholar] [CrossRef] [Green Version]
- Nederlof, R.; Eerbeek, O.; Hollmann, M.W.; Southworth, R.; Zuurbier, C.J. Targeting hexokinase II to mitochondria to modulate energy metabolism and reduce ischaemia-reperfusion injury in heart. Br. J. Pharmacol. 2014, 171, 2067–2079. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.-h.; Qiu, M.-h.; Wang, X.-j.; Sun, K.; Zheng, Y.; Jing, Z.-c. Up-regulation of hexokinase1 in the right ventricle of monocrotaline induced pulmonary hypertension. Respir. Res. 2014, 15, 119. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Wang, Q.; Liu, L.; Geng, T.; Gong, D. Mitochondrial-bound hexokinase 1 can affect the glucolipid metabolism and reactive oxygen species production in goose fatty liver. Ital. J. Anim. Sci. 2022, 21, 314–323. [Google Scholar] [CrossRef]
- Flachsbart, F.; Ufer, M.; Kleindorp, R.; Nikolaus, S.; Schreiber, S.; Nebel, A. Genetic variation in the CYP2C monooxygenase enzyme subfamily shows no association with longevity in a German population. J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2011, 66, 1186–1191. [Google Scholar] [CrossRef] [Green Version]
- Wendel, M.; Heller, A.R. Anticancer actions of omega-3 fatty acids-current state and future perspectives. Anti-Cancer Agents Med. Chem. (Former. Curr. Med. Chem.-Anti-Cancer Agents) 2009, 9, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Pearsall, E.A.; Cheng, R.; Zhou, K.; Takahashi, Y.; Matlock, H.G.; Vadvalkar, S.S.; Shin, Y.; Fredrick, T.W.; Gantner, M.L.; Meng, S. PPARα is essential for retinal lipid metabolism and neuronal survival. BMC Biol. 2017, 15, 113. [Google Scholar] [CrossRef] [PubMed]
- Steneberg, P.; Rubins, N.; Bartoov-Shifman, R.; Walker, M.D.; Edlund, H. The FFA receptor GPR40 links hyperinsulinemia, hepatic steatosis, and impaired glucose homeostasis in mouse. Cell Metab. 2005, 1, 245–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefebvre, P.; Chinetti, G.; Fruchart, J.-C.; Staels, B. Sorting out the roles of PPARα in energy metabolism and vascular homeostasis. J. Clin. Investig. 2006, 116, 571–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finck, B.N.; Lehman, J.J.; Leone, T.C.; Welch, M.J.; Bennett, M.J.; Kovacs, A.; Han, X.; Gross, R.W.; Kozak, R.; Lopaschuk, G.D. The cardiac phenotype induced by PPARα overexpression mimics that caused by diabetes mellitus. J. Clin. Investig. 2002, 109, 121–130. [Google Scholar] [CrossRef]
- Kalucka, J.; Bierhansl, L.; Conchinha, N.V.; Missiaen, R.; Elia, I.; Brüning, U.; Scheinok, S.; Treps, L.; Cantelmo, A.R.; Dubois, C. Quiescent endothelial cells upregulate fatty acid β-oxidation for vasculoprotection via redox homeostasis. Cell Metab. 2018, 28, 881–894.e13. [Google Scholar] [CrossRef] [Green Version]
- Patella, F.; Schug, Z.T.; Persi, E.; Neilson, L.J.; Erami, Z.; Avanzato, D.; Maione, F.; Hernandez-Fernaud, J.R.; Mackay, G.; Zheng, L. Proteomics-Based Metabolic Modeling Reveals That Fatty Acid Oxidation (FAO) Controls Endothelial Cell (EC) Permeability. Mol. Cell. Proteom. 2015, 14, 621–634. [Google Scholar] [CrossRef] [Green Version]
- Manicam, C.; Ginter, N.; Li, H.; Xia, N.; Goloborodko, E.; Zadeh, J.K.; Musayeva, A.; Pfeiffer, N.; Gericke, A. Compensatory vasodilator mechanisms in the ophthalmic artery of endothelial nitric oxide synthase gene knockout mice. Sci. Rep. 2017, 7, 7111. [Google Scholar] [CrossRef] [Green Version]
- Goumans, M.-J.; Liu, Z.; Ten Dijke, P. TGF-β signaling in vascular biology and dysfunction. Cell Res. 2009, 19, 116–127. [Google Scholar] [CrossRef] [Green Version]
- Ding, R.; Darland, D.C.; Parmacek, M.S.; D’amore, P.A. Endothelial–mesenchymal interactions in vitro reveal molecular mechanisms of smooth muscle/pericyte differentiation. Stem Cells Dev. 2004, 13, 509–520. [Google Scholar] [CrossRef]
- Gordon, K.J.; Blobe, G.C. Role of transforming growth factor-β superfamily signaling pathways in human disease. Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2008, 1782, 197–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Wallerath, T.; Förstermann, U. Physiological mechanisms regulating the expression of endothelial-type NO synthase. Nitric Oxide 2002, 7, 132–147. [Google Scholar] [CrossRef]
- Ten Dijke, P.; Arthur, H.M. Extracellular control of TGFβ signalling in vascular development and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Walshe, T.E.; Saint-Geniez, M.; Maharaj, A.S.; Sekiyama, E.; Maldonado, A.E.; D’Amore, P.A. TGF-β is required for vascular barrier function, endothelial survival and homeostasis of the adult microvasculature. PLoS ONE 2009, 4, e5149. [Google Scholar] [CrossRef] [Green Version]
- Rey, S.; Semenza, G.L. Hypoxia-inducible factor-1-dependent mechanisms of vascularization and vascular remodelling. Cardiovasc. Res. 2010, 86, 236–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manalo, D.J.; Rowan, A.; Lavoie, T.; Natarajan, L.; Kelly, B.D.; Ye, S.Q.; Garcia, J.G.; Semenza, G.L. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 2005, 105, 659–669. [Google Scholar] [CrossRef]
- Geraci, M.W.; Gao, B.; Shepherd, D.C.; Moore, M.D.; Westcott, J.Y.; Fagan, K.A.; Alger, L.A.; Tuder, R.M.; Voelkel, N.F. Pulmonary prostacyclin synthase overexpression in transgenic mice protects against development of hypoxic pulmonary hypertension. J. Clin. Investig. 1999, 103, 1509–1515. [Google Scholar] [CrossRef] [Green Version]
- McCullough, K.T.; Boye, S.L.; Fajardo, D.; Calabro, K.; Peterson, J.J.; Strang, C.E.; Chakraborty, D.; Gloskowski, S.; Haskett, S.; Samuelsson, S. Somatic gene editing of GUCY2D by AAV-CRISPR/Cas9 alters retinal structure and function in mouse and macaque. Hum. Gene Ther. 2019, 30, 571–589. [Google Scholar] [CrossRef]
- Mihelec, M.; Pearson, R.A.; Robbie, S.J.; Buch, P.K.; Azam, S.A.; Bainbridge, J.W.; Smith, A.J.; Ali, R.R. Long-term preservation of cones and improvement in visual function following gene therapy in a mouse model of leber congenital amaurosis caused by guanylate cyclase-1 deficiency. Hum. Gene Ther. 2011, 22, 1179–1190. [Google Scholar] [CrossRef] [PubMed]
- Lowe, D.G.; Dizhoor, A.M.; Liu, K.; Gu, Q.; Spencer, M.; Laura, R.; Lu, L.; Hurley, J.B. Cloning and expression of a second photoreceptor-specific membrane retina guanylyl cyclase (RetGC), RetGC-2. Proc. Natl. Acad. Sci. USA 1995, 92, 5535–5539. [Google Scholar] [CrossRef] [Green Version]
- Baehr, W.; Karan, S.; Maeda, T.; Luo, D.-G.; Li, S.; Bronson, J.D.; Watt, C.B.; Yau, K.-W.; Frederick, J.M.; Palczewski, K. The function of guanylate cyclase 1 and guanylate cyclase 2 in rod and cone photoreceptors. J. Biol. Chem. 2007, 282, 8837–8847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakowski, J.; Baron, M.; Bainbridge, J.; Barber, A.; Pearson, R.; Ali, R.; Sowden, J. Cone and rod photoreceptor transplantation in models of the childhood retinopathy Leber congenital amaurosis using flow-sorted Crx-positive donor cells. Hum. Mol. Genet. 2010, 19, 4545–4559. [Google Scholar] [CrossRef] [PubMed]
- Beetz, N.; Hein, L. The physiological roles of phosducin: From retinal function to stress-dependent hypertension. Cell. Mol. Life Sci. 2011, 68, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.L.; Molday, R.S. Changes in gene expression associated with retinal degeneration in the rd3 mouse. Mol. Vis. 2013, 19, 955. [Google Scholar]
- Georgiou, T.; Prokopiou, E. The new era of omega-3 fatty acids supplementation: Therapeutic effects on dry age-related macular degeneration. J. Stem Cells 2015, 10, 205–215. [Google Scholar]
- Böhm, M.R.; Pfrommer, S.; Chiwitt, C.; Brückner, M.; Melkonyan, H.; Thanos, S. Crystallin-β-b2-overexpressing NPCs support the survival of injured retinal ganglion cells and photoreceptors in rats. Investig. Ophthalmol. Vis. Sci. 2012, 53, 8265–8279. [Google Scholar] [CrossRef] [Green Version]
- Liedtke, T.; Schwamborn, J.C.; Schröer, U.; Thanos, S. Elongation of axons during regeneration involves retinal crystallin β b2 (crybb2). Mol. Cell. Proteom. 2007, 6, 895–907. [Google Scholar] [CrossRef] [Green Version]
- Aryal, S.; Hussain, S.; Drevon, C.A.; Nagelhus, E.; Hvalby, Ø.; Jensen, V.; Walaas, S.I.; Davanger, S. Omega-3 fatty acids regulate plasticity in distinct hippocampal glutamatergic synapses. Eur. J. Neurosci. 2019, 49, 40–50. [Google Scholar] [CrossRef]
- Hajjar, T.; Goh, Y.M.; Rajion, M.A.; Vidyadaran, S.; Li, T.A.; Ebrahimi, M. Alterations in neuronal morphology and synaptophysin expression in the rat brain as a result of changes in dietary n-6: N-3 fatty acid ratios. Lipids Health Dis. 2013, 12, 113. [Google Scholar] [CrossRef] [Green Version]
- Su, H.-M. Mechanisms of n-3 fatty acid-mediated development and maintenance of learning memory performance. J. Nutr. Biochem. 2010, 21, 364–373. [Google Scholar] [CrossRef]
- Dan, C.; Jian-Bin, T.; Hui, W.; Le-Ping, Z.; Jin, Z.; Ju-Fang, H.; Xue-Gang, L. Synaptophysin expression in rat retina following acute high intraocular pressure. Acta Histochem. Cytochem. 2008, 41, 173–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joyal, J.-S.; Gantner, M.L.; Smith, L.E. Retinal energy demands control vascular supply of the retina in development and disease: The role of neuronal lipid and glucose metabolism. Prog. Retin. Eye Res. 2018, 64, 131. [Google Scholar] [CrossRef] [PubMed]
- Gehrig, A.; Langmann, T.; Horling, F.; Janssen, A.; Bonin, M.; Walter, M.; Poths, S.; Weber, B.H. Genome-wide expression profiling of the retinoschisin-deficient retina in early postnatal mouse development. Investig. Ophthalmol. Vis. Sci. 2007, 48, 891–900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kjellström, S.; Ghosh, F.; Vijayasarathy, C.; Andréasson, S. Alteration of Vitreal Retinoschisin Level in Human Primary Retinal Detachment. JAMA Ophthalmol. 2014, 132, 353–354. [Google Scholar] [CrossRef]
- Bianchi, S.; van Riel, W.E.; Kraatz, S.H.; Olieric, N.; Frey, D.; Katrukha, E.A.; Jaussi, R.; Missimer, J.; Grigoriev, I.; Olieric, V. Structural basis for misregulation of kinesin KIF21A autoinhibition by CFEOM1 disease mutations. Sci. Rep. 2016, 6, 30668. [Google Scholar] [CrossRef]
- Morfini, G.A.; Burns, M.; Binder, L.I.; Kanaan, N.M.; LaPointe, N.; Bosco, D.A.; Brown, R.H.; Brown, H.; Tiwari, A.; Hayward, L. Axonal transport defects in neurodegenerative diseases. J. Neurosci. 2009, 29, 12776–12786. [Google Scholar] [CrossRef] [Green Version]
- Vivian, A.J. Congenital fibrosis of the extra-ocular muscles (CFEOM) and the cranial dysinnervation disorders. Eye 2020, 34, 251–255. [Google Scholar] [CrossRef]
- Sharoukhov, D.; Bucinca-Cupallari, F.; Lim, H. Microtubule imaging reveals cytoskeletal deficit predisposing the retinal ganglion cell axons to atrophy in DBA/2J. Investig. Ophthalmol. Vis. Sci. 2018, 59, 5292–5300. [Google Scholar] [CrossRef] [Green Version]
- Sherratt, S.C.; Dawoud, H.; Bhatt, D.L.; Malinski, T.; Mason, R.P. Omega-3 and omega-6 fatty acids have distinct effects on endothelial fatty acid content and nitric oxide bioavailability. Prostaglandins Leukot. Essent. Fat. Acids 2021, 173, 102337. [Google Scholar] [CrossRef]
- Martins, M.A.; Moss, M.B.; Mendes, I.K.; Águila, M.B.; Mandarim-de-Lacerda, C.A.; Brunini, T.M.; Mendes-Ribeiro, A.C. Role of dietary fish oil on nitric oxide synthase activity and oxidative status in mice red blood cells. Food Funct. 2014, 5, 3208–3215. [Google Scholar] [CrossRef]
- Felau, S.M.; Sales, L.P.; Solis, M.Y.; Hayashi, A.P.; Roschel, H.; Sá-Pinto, A.L. Omega-3 fatty acid supplementation improves endothelial function in primary antiphospholipid syndrome: A small-scale randomized double-blind placebo-controlled trial. Front. Immunol. 2018, 9, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stahl, A.; Sapieha, P.; Connor, K.M.; SanGiovanni, J.P.; Chen, J.; Aderman, C.M. PPARγ mediates a direct antiangiogenic effect of ω3-PUFAs in proliferative retinopathy. Circ. Res. 2010, 107, 495–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delbosc, S.; Glorian, M.; Le Port, A.-S.; Béréziat, G.; Andréani, M.; Limon, I. The benefit of docosahexanoic acid on the migration of vascular smooth muscle cells is partially dependent on Notch regulation of MMP-2/-9. Am. J. Pathol. 2008, 172, 1430–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Bae, M.; Park, Y.-K.; Lee, J.-Y. n-3 PUFAs inhibit TGFβ1-induced profibrogenic gene expression by ameliorating the repression of PPARγ in hepatic stellate cells. J. Nutr. Biochem. 2020, 85, 108452. [Google Scholar] [CrossRef]
- Holla, V.R.; Adas, F.; Imig, J.D.; Zhao, X.; Price, E., Jr.; Olsen, N. Alterations in the regulation of androgen-sensitive Cyp 4a monooxygenases cause hypertension. Proc. Natl. Acad. Sci. USA 2001, 98, 5211–5216. [Google Scholar] [CrossRef] [Green Version]
- Holmdahl, R.; Malissen, B. The Need for Littermate Controls; Wiley Online Library: Hoboken, NJ, USA, 2012; pp. 45–47. [Google Scholar]
- Jiménez, J.A.; Zylka, M.J. Controlling litter effects to enhance rigor and reproducibility with rodent models of neurodevelopmental disorders. J. Neurodev. Disord. 2021, 13, 1–9. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perumal, N.; Herfurth, A.; Pfeiffer, N.; Manicam, C. Short-Term Omega-3 Supplementation Modulates Novel Neurovascular and Fatty Acid Metabolic Proteome Changes in the Retina and Ophthalmic Artery of Mice with Targeted Cyp2c44 Gene Deletion. Cells 2022, 11, 3494. https://doi.org/10.3390/cells11213494
Perumal N, Herfurth A, Pfeiffer N, Manicam C. Short-Term Omega-3 Supplementation Modulates Novel Neurovascular and Fatty Acid Metabolic Proteome Changes in the Retina and Ophthalmic Artery of Mice with Targeted Cyp2c44 Gene Deletion. Cells. 2022; 11(21):3494. https://doi.org/10.3390/cells11213494
Chicago/Turabian StylePerumal, Natarajan, Anna Herfurth, Norbert Pfeiffer, and Caroline Manicam. 2022. "Short-Term Omega-3 Supplementation Modulates Novel Neurovascular and Fatty Acid Metabolic Proteome Changes in the Retina and Ophthalmic Artery of Mice with Targeted Cyp2c44 Gene Deletion" Cells 11, no. 21: 3494. https://doi.org/10.3390/cells11213494
APA StylePerumal, N., Herfurth, A., Pfeiffer, N., & Manicam, C. (2022). Short-Term Omega-3 Supplementation Modulates Novel Neurovascular and Fatty Acid Metabolic Proteome Changes in the Retina and Ophthalmic Artery of Mice with Targeted Cyp2c44 Gene Deletion. Cells, 11(21), 3494. https://doi.org/10.3390/cells11213494