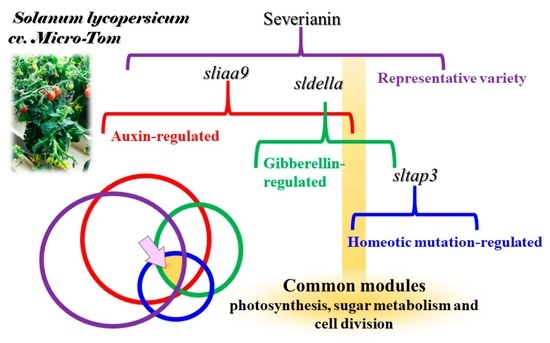
Transcriptomic, Hormonomic and Metabolomic Analyses Highlighted the Common Modules Related to Photosynthesis, Sugar Metabolism and Cell Division in Parthenocarpic Tomato Fruits during Early Fruit Set
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Microarray Analysis
2.3. Metabolomic Profiling Analysis
2.4. RNA-Seq Analysis
2.5. Weighted Correlation Network Analysis (WCNA) of Transcriptome and Metabolome Datasets
2.6. Data Analysis
3. Results
3.1. Comparison of Ovary/Fruit Development of WT and the Mutants during Early Fruit Set
3.2. Plant Hormone Accumulation Patterns during Fruit Set in WT and the Mutants with and without Pollination
3.3. General Trend of the Transcriptomic and Metabolomic Changes of the Mutants
3.4. WCNA of the Transcript and Metabolite Profiles of the Mutants
3.5. Transcript Changes in Photosynthesis, Sugar Metabolism and Cell Wall Biosynthesis during Tomato Fruit Set
3.6. Metabolite Correlations in Photosynthesis and Carbohydrate Metabolism and Extracted by Network Analysis
3.7. RNA-Seq Analysis of a Parthenocarpic Cultivar ‘Severianin’ Reveals the Similar Trends of Transcript Changes Found throughout the Three Micro-Tom Mutants
4. Discussion
4.1. Endogenous Accumulation of IAA and Active GAs Has no Relation for Induction of Parthenocarpy of the Mutants Lacking SlIAA9, SlDELLA and SlTAP3
4.2. Photosynthesis, Sugar Metabolism and Cell Wall Pathways Are Potential Candidates Contributing Parthenocarpy in the Mutant Tomato
4.3. The Proof-of-Concept Approach Could Objectively Extract the Common Genes with Similar Behaviour in Parthenocarpic Tomato
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ficcadenti, N.; Sestili, S.; Pandolfini, T.; Cirillo, C.; Rotino, G.L.; Spena, A. Genetic engineering of parthenocarpic fruit development in tomato. Mol. Breed. 1999, 5, 463–470. [Google Scholar] [CrossRef] [Green Version]
- Acciarri, N.; Restaino, F.; Vitelli, G.; Perrone, D.; Zottini, M.; Pandolfini, T.; Spena, A.; Rotino, G.L. Genetically modified parthenocarpic eggplants: Improved fruit productivity under both greenhouse and open field cultivation. BMC Biotechnol. 2002, 2, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serrani, J.C.; Fos, M.; Atarés, A.; García-Martínez, J.L. Effect of gibberellin and auxin on parthenocarpic fruit growth induction in the cv Micro-Tom of tomato. J. Plant Growth Regul. 2007, 26, 211–221. [Google Scholar] [CrossRef]
- De Jong, M.; Mariani, C.; Vriezen, W.H. The role of auxin and gibberellin in tomato fruit set. J. Exp. Bot. 2009, 60, 1523–1532. [Google Scholar] [CrossRef] [Green Version]
- Gorguet, B.; Van Heusden, A.W.; Lindhout, P. Parthenocarpic fruit development in tomato. Plant Biol. 2005, 7, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Blanvillain, R.; Delseny, M.; Gallois, P.; Varoquaux, F. Less is better: New approaches for seedless fruit production. Trends Biotechnol. 2000, 18, 233–242. [Google Scholar] [CrossRef]
- Serrani, J.C.; Ruiz-Rivero, O.; Fos, M.; García-Martínez, J.L. Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J. 2008, 56, 922–934. [Google Scholar] [CrossRef] [Green Version]
- Schijlen, E.G.W.M.; De Vos, C.H.R.; Martens, S.; Jonker, H.H.; Rosin, F.M.; Molthoff, J.W.; Tikunov, Y.M.; Angenent, G.C.; Van Tunen, A.J.; Bovy, A.G. RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol. 2007, 144, 1520–1530. [Google Scholar] [CrossRef] [Green Version]
- Molesini, B.; Pandolfini, T.; Rotino, G.L.; Dani, V.; Spena, A. Aucsia gene silencing causes parthenocarpic fruit development in tomato. Plant Physiol. 2009, 149, 534–548. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Gracia, P.; Roque, E.; Medina, M.; Rochina, M.; Hamza, R.; Angarita-Díaz, M.P.; Moreno, V.; Pérez-Martín, F.; Lozano, R.; Cañas, L.; et al. The parthenocarpic hydra mutant reveals a new function for a SPOROCYTELESS-like gene in the control of fruit set in tomato. New Phytol. 2017, 214, 1198–1212. [Google Scholar] [CrossRef] [Green Version]
- Gustafson, F.G. The cause of natural parthenocarpy. Am. J. Bot. 1939, 26, 135–138. [Google Scholar] [CrossRef]
- Hu, J.; Israeli, A.; Ori, N.; Sun, T.P. The interaction between DELLA and ARF/IAA mediates crosstalk between gibberellin and auxin signaling to control fruit initiation in tomato. Plant Cell 2018, 30, 1710–1728. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, A.; Handa, A.K. Hormonal regulation of tomato fruit development: A molecular perspective. J. Plant Growth Regul. 2005, 24, 67–82. [Google Scholar] [CrossRef] [Green Version]
- Bassel, G.W.; Mullen, R.T.; Bewley, J.D. Procera is a putative DELLA mutant in tomato (Solanum lycopersicum): Effects on the seed and vegetative plant. J. Exp. Bot. 2008, 59, 585–593. [Google Scholar] [CrossRef] [Green Version]
- Audran-Delalande, C.; Bassa, C.; Mila, I.; Regad, F.; Zouine, M.; Bouzayen, M. Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in Tomato. Plant Cell Physiol. 2012, 53, 659–672. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Jones, B.; Li, Z.; Frasse, P.; Delalande, C.; Regad, F.; Chaabouni, S.; Latché, A.; Pech, J.C.; Bouzayen, M. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 2005, 17, 2676–2692. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, H.; Hirano, K.; Sato, T.; Mitsuda, N.; Nomoto, M.; Maeo, K.; Koketsu, E.; Mitani, R.; Kawamura, M.; Ishiguro, S.; et al. DELLA protein functions as a transcriptional activator through the DNA binding of the INDETERMINATE DOMAIN family proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 7861–7866. [Google Scholar] [CrossRef] [Green Version]
- Carrera, E.; Ruiz-Rivero, O.; Peres, L.E.P.; Atares, A.; Garcia-Martinez, J.L. Characterization of the procera tomato mutant shows novel functions of the SlDELLA protein in the control of flower morphology, cell division and expansion, and the auxin-signaling pathway during fruit-set and development. Plant Physiol. 2012, 160, 1581–1596. [Google Scholar] [CrossRef] [Green Version]
- Martí, C.; Orzáez, D.; Ellul, P.; Moreno, V.; Carbonell, J.; Granell, A. Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 2007, 52, 865–876. [Google Scholar] [CrossRef]
- Livne, S.; Lor, V.S.; Nir, I.; Eliaz, N.; Aharoni, A.; Olszewski, N.E.; Eshed, Y.; Weiss, D. Uncovering DELLA-independent gibberellin responses by characterizing new tomato procera mutants. Plant Cell 2015, 27, 1579–1594. [Google Scholar] [CrossRef] [Green Version]
- Honma, T.; Goto, K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001, 409, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATTA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef] [PubMed]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Schuch, W.; Giovannoni, J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Dusi, V.; Pennisi, F.; Pandolfini, T. How hormones and mads-box transcription factors are involved in controlling fruit set and parthenocarpy in tomato. Genes 2020, 11, 1441. [Google Scholar] [CrossRef] [PubMed]
- Ampomah-Dwamena, C.; Morris, B.A.; Sutherland, P.; Veit, B.; Yao, J.L. Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol. 2002, 130, 605–617. [Google Scholar] [CrossRef] [Green Version]
- De Martino, G.; Pan, I.; Emmanuel, E.; Levy, A.; Irish, V.F. Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development. Plant Cell 2006, 18, 1833–1845. [Google Scholar] [CrossRef] [Green Version]
- Okabe, Y.; Yamaoka, T.; Ariizumi, T.; Ushijima, K.; Kojima, M.; Takebayashi, Y.; Sakakibara, H.; Kusano, M.; Shinozaki, Y.; Pulungan, S.I.; et al. Aberrant Stamen Development is Associated with Parthenocarpic Fruit Set Through Up-Regulation of Gibberellin Biosynthesis in Tomato. Plant Cell Physiol. 2019, 60, 38–51. [Google Scholar] [CrossRef]
- Vardy, E.; Lapushner, D.; Genizi, A.; Hewitt, J. Genetics of parthenocarpy in tomato under a low temperature regime: II Cultivar ‘Severianin’. Euphytica 1989, 41, 9–15. [Google Scholar] [CrossRef]
- Lin, S.; Splittstoesser, W.E.; George, W.L. Factors controlling the expression of parthenocarpy in ‘Severianin’ tomato. Sci. Hortic. 1983, 19, 45–53. [Google Scholar] [CrossRef]
- Lin, S.; George, W.L.; Splittstoesser, W.E. Expression and inheritance of parthenocarpy in ‘severianin’ tomato. J. Hered. 1984, 75, 62–66. [Google Scholar] [CrossRef]
- Fos, M.; Nuez, F.; García-Martínez, J.L. The gene pat-2, which induces natural parthenocarpy, alters the gibberellin content in unpollinated tomato ovaries. Plant Physiol. 2000, 122, 471–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbona, V.; Manzi, M.; de Ollas, C.; Gómez-Cadenas, A. Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Saito, K. Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef] [Green Version]
- Flis, A.; Mengin, V.; Ivakov, A.A.; Mugford, S.T.; Hubberten, H.M.; Encke, B.; Krohn, N.; Höhne, M.; Feil, R.; Hoefgen, R.; et al. Multiple circadian clock outputs regulate diel turnover of carbon and nitrogen reserves. Plant Cell Environ. 2019, 42, 549–573. [Google Scholar] [CrossRef]
- Kusano, M.; Fukushima, A.; Tabuchi-Kobayashi, M.; Funayama, K.; Kojima, S.; Maruyama, K.; Yamamoto, Y.Y.; Nishizawa, T.; Kobayashi, M.; Wakazaki, M.; et al. Cytosolic glutamine synthetase1;1 modulates metabolism and chloroplast development in ROOTs1[open]. Plant Physiol. 2020, 182, 1894–1909. [Google Scholar] [CrossRef] [Green Version]
- Kojima, M.; Kamada-Nobusada, T.; Komatsu, H.; Takei, K.; Kuroha, T.; Mizutani, M.; Ashikari, M.; Ueguchi-Tanaka, M.; Matsuoka, M.; Suzuki, K.; et al. Highly sensitive and high-throughput analysis of plant hormones using ms-probe modification and liquid chromatographytandem mass spectrometry: An application for hormone profiling in oryza sativa. Plant Cell Physiol. 2009, 50, 1201–1214. [Google Scholar] [CrossRef]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef] [Green Version]
- Todaka, D.; Zhao, Y.; Yoshida, T.; Kudo, M.; Kidokoro, S.; Mizoi, J.; Kodaira, K.S.; Takebayashi, Y.; Kojima, M.; Sakakibara, H.; et al. Temporal and spatial changes in gene expression, metabolite accumulation and phytohormone content in rice seedlings grown under drought stress conditions. Plant J. 2017, 90, 61–78. [Google Scholar] [CrossRef]
- Fukushima, A.; Kuroha, T.; Nagai, K.; Hattori, Y.; Kobayashi, M.; Nishizawa, T.; Kojima, M.; Utsumi, Y.; Oikawa, A.; Seki, M.; et al. Metabolite and phytohormone profiling illustrates metabolic reprogramming as an escape strategy of deepwater rice during partially submerged stress. Metabolites 2020, 10, 68. [Google Scholar] [CrossRef] [Green Version]
- Urano, K.; Maruyama, K.; Ogata, Y.; Morishita, Y.; Takeda, M.; Sakurai, N.; Suzuki, H.; Saito, K.; Shibata, D.; Kobayashi, M.; et al. Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 2009, 57, 1065–1078. [Google Scholar] [CrossRef]
- Diretto, G.; Al-Babili, S.; Tavazza, R.; Scossa, F.; Papacchioli, V.; Migliore, M.; Beyer, P.; Giuliano, G. Transcriptional-metabolic networks in β-carotene-enriched potato tubers: The long and winding road to the golden phenotype. Plant Physiol. 2010, 154, 899–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osorio, S.; Alba, R.; Nikoloski, Z.; Kochevenko, A.; Fernie, A.R.; Giovannoni, J.J. Integrative comparative analyses of transcript and metabolite profiles from pepper and tomato ripening and development stages uncovers species-specific patterns of network regulatory behavior. Plant Physiol. 2012, 159, 1713–1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichihashi, Y.; Date, Y.; Shino, A.; Shimizu, T.; Shibata, A.; Kumaishi, K.; Funahashi, F.; Wakayama, K.; Yamazaki, K.; Umezawa, A.; et al. Multi-omics analysis on an agroecosystem reveals the significant role of organic nitrogen to increase agricultural crop yield. Proc. Natl. Acad. Sci. USA 2020, 117, 14552–14560. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Li, R.; Weng, L.; Sun, Y.; Li, M.; Xiao, H. Domain-specific expression of meristematic genes is defined by the LITTLE ZIPPER protein DTM in tomato. Commun. Biol. 2019, 2, 134. [Google Scholar] [CrossRef] [Green Version]
- Shinozaki, Y.; Beauvoit, B.P.; Takahara, M.; Hao, S.; Ezura, K.; Andrieu, M.H.; Nishida, K.; Mori, K.; Suzuki, Y.; Kuhara, S.; et al. Fruit setting rewires central metabolism via gibberellin cascades. Proc. Natl. Acad. Sci. USA 2020, 117, 23970–23981. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Schauer, N.; Usadel, B.; Frasse, P.; Zouine, M.; Hernould, M.; Latché, A.; Pech, J.C.; Fernie, A.R.; Bouzayena, M. Regulatory features underlying pollination-dependent and-independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 2009, 21, 1428–1452. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.L.; Gross, K.C. A family of at least seven β-galactosidase genes is expressed during tomato fruit development. Plant Physiol. 2000, 123, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, S.; Ljung, K.; Sorefan, K.; Alvey, E.; Harberd, N.P.; Østergaard, L. Fruit growth in Arabidopsis occurs via DELLA-dependent and DELLA-independent gibberellin responses. Plant Cell 2012, 24, 3982–3996. [Google Scholar] [CrossRef] [Green Version]
- Bu, H.; Yu, W.; Yuan, H.; Yue, P.; Wei, Y.; Wang, A. Endogenous auxin content contributes to larger size of apple Fruit. Front. Plant Sci. 2020, 11, 592540. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, L.; Picciarelli, P.; Lombardi, L.; Ceccarelli, N. Fruit-set and early fruit growth in tomato are associated with increases in indoleacetic acid, cytokinin, and bioactive gibberellin contents. J. Plant Growth Regul. 2011, 30, 405–415. [Google Scholar] [CrossRef]
- García-Hurtado, N.; Carrera, E.; Ruiz-Rivero, O.; López-Gresa, M.P.; Hedden, P.; Gong, F.; García-Martínez, J.L. The characterization of transgenic tomato overexpressing gibberellin 20-oxidase reveals induction of parthenocarpic fruit growth, higher yield, and alteration of the gibberellin biosynthetic pathway. J. Exp. Bot. 2012, 63, 5803–5813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, L.; Yue, R.; Wang, H.; Liu, J.; Zhai, R.; Yang, J.; Wu, M.; Si, M.; Zhang, H.; Yang, C.; et al. 2,4-D-induced parthenocarpy in pear is mediated by enhancement of GA4 biosynthesis. Physiol. Plant. 2019, 166, 812–820. [Google Scholar] [CrossRef] [PubMed]
- De Jong, M.; Wolters-Arts, M.; Schimmel, B.C.J.; Stultiens, C.L.M.; De Groot, P.F.M.; Powers, S.J.; Tikunov, Y.M.; Bovy, A.G.; Mariani, C.; Vriezen, W.H.; et al. Solanum lycopersicum AUXIN RESPONSE FACTOR 9 regulates cell division activity during early tomato fruit development. J. Exp. Bot. 2015, 66, 3405–3416. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Dinh, T.T.; Li, D.; Shi, B.; Li, Y.; Cao, X.; Guo, L.; Pan, Y.; Jiao, Y.; Chen, X. AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 2014, 80, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Gu, X.; Shao, J.; Hu, Z.; Yang, W.; Wang, L.; Su, H.; Zhu, L. Auxin metabolism is involved in fruit set and early fruit development in the parthenocarpic tomato “R35-P”. Front. Plant Sci. 2021, 12, 671713. [Google Scholar] [CrossRef]
- Larkin, R.M. Tetrapyrrole signaling in plants. Front. Plant Sci. 2016, 7, 1586. [Google Scholar] [CrossRef] [Green Version]
- Masuda, T.; Fusada, N.; Oosawa, N.; Takamatsu, K.i.; Yamamoto, Y.Y.; Ohto, M.; Nakamura, K.; Goto, K.; Shibata, D.; Shirano, Y. Functional analysis of isoforms of NADPH: Protochlorophyllide oxidoreductase (POR), PORB and PORC, in Arabidopsis thaliana. Plant Cell Physiol. 2003, 44, 963–974. [Google Scholar] [CrossRef] [Green Version]
- Tang, N.; Deng, W.; Hu, G.; Hu, N.; Li, Z. Transcriptome profiling reveals the regulatory mechanism underlying pollination dependent and parthenocarpic fruit set mainly mediated by auxin and gibberellin. PLoS ONE 2015, 10, e0125355. [Google Scholar] [CrossRef] [Green Version]
- Malladi, A.; Johnson, L.K. Expression profiling of cell cycle genes reveals key facilitators of cell production during carpel development, fruit set, and fruit growth in apple (Malus×domestica Borkh.). J. Exp. Bot. 2011, 62, 205–219. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, R.G.; Johnston, S.L.; Yauk, Y.K.; Sharma, N.N.; Schröder, R. Analysis of xyloglucan endotransglucosylase/hydrolase (XTH) gene families in kiwifruit and apple. Postharvest Biol. Technol. 2009, 51, 149–157. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Diffuse growth of plant cell walls. Plant Physiol. 2018, 176, 16–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kende, H.; Bradford, K.J.; Brummell, D.A.; Cho, H.T.; Cosgrove, D.J.; Fleming, A.J.; Gehring, C.; Lee, Y.; McQueen-Mason, S.; Rose, J.K.C.; et al. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol. Biol. 2004, 55, 311–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klinghammer, M.; Tenhaken, R. Genome-wide analysis of the UDP-glucose dehydrogenase gene family in Arabidopsis, a key enzyme for matrix polysaccharides in cell walls. J. Exp. Bot. 2007, 58, 3609–3621. [Google Scholar] [CrossRef] [PubMed]
- Runge, S.; Sperling, U.; Frick, G.; Apel, K.; Armstrong, G.A. Distinct roles for light-dependent NADPH: Protochlorophyllide oxidoreductases (POR) A and B during greening in higher plants. Plant J. 1996, 9, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Weng, L.; Zhao, F.; Li, R.; Xu, C.; Chen, K.; Xiao, H. The zinc finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. Plant Physiol. 2015, 167, 931–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Odanaka, S.; Bennett, A.B.; Kanayama, Y. Distinct physiological roles of fructokinase isozymes revealed by gene-specific suppression of Frk1 and Frk2 expression in tomato. Plant Physiol. 2002, 129, 1119–1126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Zhang, Y.; Feng, Q.; Qin, L.; Pan, C.; Lamin-Samu, A.T.; Lu, G. Tomato AUXIN RESPONSE FACTOR 5 regulates fruit set and development via the mediation of auxin and gibberellin signaling. Sci. Rep. 2018, 8, 2971. [Google Scholar] [CrossRef] [Green Version]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kusano, M.; Worarad, K.; Fukushima, A.; Kamiya, K.; Mitani, Y.; Okazaki, Y.; Higashi, Y.; Nakabayashi, R.; Kobayashi, M.; Mori, T.; et al. Transcriptomic, Hormonomic and Metabolomic Analyses Highlighted the Common Modules Related to Photosynthesis, Sugar Metabolism and Cell Division in Parthenocarpic Tomato Fruits during Early Fruit Set. Cells 2022, 11, 1420. https://doi.org/10.3390/cells11091420
Kusano M, Worarad K, Fukushima A, Kamiya K, Mitani Y, Okazaki Y, Higashi Y, Nakabayashi R, Kobayashi M, Mori T, et al. Transcriptomic, Hormonomic and Metabolomic Analyses Highlighted the Common Modules Related to Photosynthesis, Sugar Metabolism and Cell Division in Parthenocarpic Tomato Fruits during Early Fruit Set. Cells. 2022; 11(9):1420. https://doi.org/10.3390/cells11091420
Chicago/Turabian StyleKusano, Miyako, Kanjana Worarad, Atsushi Fukushima, Ken Kamiya, Yuka Mitani, Yozo Okazaki, Yasuhiro Higashi, Ryo Nakabayashi, Makoto Kobayashi, Tetsuya Mori, and et al. 2022. "Transcriptomic, Hormonomic and Metabolomic Analyses Highlighted the Common Modules Related to Photosynthesis, Sugar Metabolism and Cell Division in Parthenocarpic Tomato Fruits during Early Fruit Set" Cells 11, no. 9: 1420. https://doi.org/10.3390/cells11091420
APA StyleKusano, M., Worarad, K., Fukushima, A., Kamiya, K., Mitani, Y., Okazaki, Y., Higashi, Y., Nakabayashi, R., Kobayashi, M., Mori, T., Nishizawa, T., Takebayashi, Y., Kojima, M., Sakakibara, H., Saito, K., Hao, S., Shinozaki, Y., Okabe, Y., Kimbara, J., ... Ezura, H. (2022). Transcriptomic, Hormonomic and Metabolomic Analyses Highlighted the Common Modules Related to Photosynthesis, Sugar Metabolism and Cell Division in Parthenocarpic Tomato Fruits during Early Fruit Set. Cells, 11(9), 1420. https://doi.org/10.3390/cells11091420