Plasma Sphingomyelin Disturbances: Unveiling Its Dual Role as a Crucial Immunopathological Factor and a Severity Prognostic Biomarker in COVID-19
Abstract
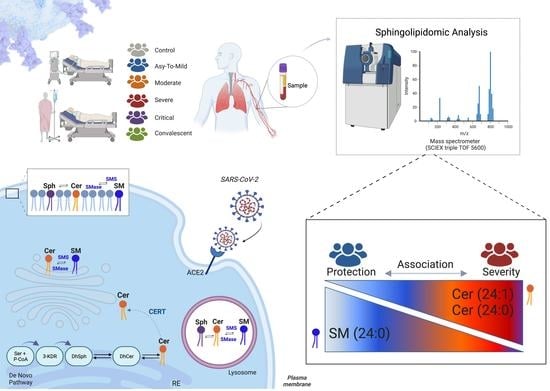
:1. Introduction
2. Material and Methods
2.1. Study Design and Blood Collection
2.2. Ethical Considerations
2.3. Laboratory and Data Collection
2.4. Cytokine Measurements
2.5. Lipid Extraction and Sample Preparation for LC-MS/MS
2.6. Sphingolipid Quantification by LC-MS/MS
2.7. RNA Extraction and Analysis
2.8. Transcriptome Profiling
2.8.1. Bioinformatic Analysis of Transcriptome Data
2.8.2. Validation of Microarray Data by Reverse Transcription Quantitative Real-Time PCR
2.9. Statistical Data Analysis
3. Results
3.1. Characterization of Study Participants
3.2. COVID-19 Severity Increased Gene Expression of Key Enzymes Involved in SM and Cer Synthesis
3.3. Plasma SM Profile Is Associated with COVID-19 and Can Be a Potential Biomarker for Assessing Severity of Disease
3.4. The Discovered Plasma SM Species Panel Effectively Distinguished Severe COVID-19
3.5. Multivariate Binomial Logistic Regression Determines the Association between Cer/SM Species and COVID-19 Clinical Severity and Mortality
3.6. Correlation of Values of SM Species with Immunological, Clinical, and Laboratory Markers in COVID-19
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Gandhi, R.T.; Lynch, J.B.; del Rio, C. Mild or Moderate COVID-19. N. Engl. J. Med. 2020, 383, 1757–1766. [Google Scholar] [CrossRef] [PubMed]
- Murthy, S.; Gomersall, C.D.; Fowler, R.A. Care for Critically Ill Patients with COVID-19. J. Am. Med. Assoc. 2020, 323, 1499–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erdinc, B.; Sahni, S.; Gotlieb, V. Hematological manifestations and complications of COVID-19. Adv. Clin. Exp. Med. 2021, 30, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; de Giglio, M.; Roviello, G. SARS-CoV-2: Recent Reports on Antiviral Therapies Based on Lopinavir/Ritonavir, Darunavir/Umifenovir, Hydroxychloroquine, Remdesivir, Favipiravir and other Drugs for the Treatment of the New Coronavirus. Curr. Med. Chem. 2020, 27, 4536–4541. [Google Scholar] [CrossRef] [PubMed]
- Carpinteiro, A.; Edwards, M.J.; Hoffmann, M.; Kochs, G.; Gripp, B.; Weigang, S.; Adams, C.; Carpinteiro, E.; Gulbins, A.; Keitsch, S.; et al. Pharmacological Inhibition of Acid Sphingomyelinase Prevents Uptake of SARS-CoV-2 by Epithelial Cells. Cell Rep. Med. 2020, 1, 100142. [Google Scholar] [CrossRef]
- Schneider-Schaulies, J.; Schneider-Schaulies, S. Sphingolipids in viral infection. Biol. Chem. 2015, 396, 585–595. [Google Scholar] [CrossRef]
- Simonis, A.; Schubert-Unkmeir, A. The role of acid sphingomyelinase and modulation of sphingolipid metabolism in bacterial infection. Biol. Chem. 2018, 399, 1135–1146. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 175–191. [Google Scholar] [CrossRef]
- Gomez-Larrauri, A.; Presa, N.; Dominguez-Herrera, A.; Ouro, A.; Trueba, M.; Gomez-Muñoz, A. Role of bioactive sphingolipids in physiology and pathology. Essays Biochem. 2020, 64, 579–589. [Google Scholar] [CrossRef]
- Hussain, G.; Wang, J.; Rasul, A.; Anwar, H.; Imran, A.; Qasim, M.; Zafar, S.; Kamran, S.K.S.; Razzaq, A.; Aziz, N.; et al. Role of cholesterol and sphingolipids in brain development and neurological diseases. Lipids Health Dis. 2019, 18, 26. [Google Scholar] [CrossRef] [Green Version]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef] [Green Version]
- Vitner, E.B.; Achdout, H.; Avraham, R.; Politi, B.; Cherry, L.; Tamir, H.; Yahalom-Ronen, Y.; Paran, N.; Melamed, S.; Erez, N.; et al. Glucosylceramide synthase inhibitors prevent replication of SARS-CoV-2 and influenza virus. J. Biol. Chem. 2021, 296, 100470. [Google Scholar] [CrossRef]
- Bezgovsek, J.; Gulbins, E.; Friedrich, S.-K.; Lang, K.S.; Duhan, V. Sphingolipids in early viral replication and innate immune activation. Biol. Chem. 2018, 399, 1115–1123. [Google Scholar] [CrossRef]
- Wigger, D.; Gulbins, E.; Kleuser, B.; Schumacher, F. Monitoring the Sphingolipid de novo Synthesis by Stable-Isotope Labeling and Liquid Chromatography-Mass Spectrometry. Front. Cell Dev. Biol. 2019, 7, 210. [Google Scholar] [CrossRef] [Green Version]
- Kurz, J.; Parnham, M.J.; Geisslinger, G.; Schiffmann, S. Ceramides as Novel Disease Biomarkers. Trends Mol. Med. 2019, 25, 20–32. [Google Scholar] [CrossRef]
- Gandy, K.A.O.; Obeid, L.M. Regulation of the Sphingosine Kinase/Sphingosine 1-Phosphate Pathway BT—Sphingolipids in Disease; Gulbins, E., Petrache, I., Eds.; Springer: Vienna, Austria, 2013; pp. 275–303. [Google Scholar] [CrossRef]
- Pyne, S.; Adams, D.R.; Pyne, N.J. Sphingosine 1-phosphate and sphingosine kinases in health and disease: Recent advances. Prog. Lipid Res. 2016, 62, 93–106. [Google Scholar] [CrossRef] [Green Version]
- Törnquist, K.; Asghar, M.Y.; Srinivasan, V.; Korhonen, L.; Lindholm, D. Sphingolipids as Modulators of SARS-CoV-2 Infection. Front. Cell Dev. Biol. 2021, 9, 689854. [Google Scholar] [CrossRef]
- Vitner, E.B.; Avraham, R.; Politi, B.; Melamed, S.; Israely, T. Elevation in sphingolipid upon SARS-CoV-2 infection: Possible implications for COVID-19 pathology. Life Sci. Alliance 2022, 5, e202101168. [Google Scholar] [CrossRef]
- Marín-Corral, J.; Rodríguez-Morató, J.; Gomez-Gomez, A.; Pascual-Guardia, S.; Muñoz-Bermúdez, R.; Salazar-Degracia, A.; Pérez-Terán, P.; Restrepo, M.I.; Khymenets, O.; Haro, N.; et al. Metabolic Signatures Associated with Severity in Hospitalized COVID-19 Patients. Int. J. Mol. Sci. 2021, 22, 4794. [Google Scholar] [CrossRef]
- Lee, J.W.; Su, Y.; Baloni, P.; Chen, D.; Pavlovitch-Bedzyk, A.J.; Yuan, D.; Duvvuri, V.R.; Ng, R.H.; Choi, J.; Xie, J.; et al. Integrated analysis of plasma and single immune cells uncovers metabolic changes in individuals with COVID-19. Nat. Biotechnol. 2022, 40, 110–120. [Google Scholar] [CrossRef]
- Torretta, E.; Garziano, M.; Poliseno, M.; Capitanio, D.; Biasin, M.; Santantonio, T.A.; Clerici, M.; Lo Caputo, S.; Trabattoni, D.; Gelfi, C. Severity of COVID-19 Patients Predicted by Serum Sphingolipids Signature. Int. J. Mol. Sci. 2021, 22, 10198. [Google Scholar] [CrossRef] [PubMed]
- Janneh, A.H.; Kassir, M.F.; Dwyer, C.J.; Chakraborty, P.; Pierce, J.S.; Flume, P.A.; Li, H.; Nadig, S.N.; Mehrotra, S.; Ogretmen, B. Alterations of lipid metabolism provide serologic biomarkers for the detection of asymptomatic versus symptomatic COVID-19 patients. Sci. Rep. 2021, 11, 14232. [Google Scholar] [CrossRef] [PubMed]
- Diagnosis and treatment protocol for novel coronavirus pneumonia (Trial version 7). Chin. Med. J. 2020, 133, 1087–1095. [CrossRef] [PubMed]
- Wan, S.; Xiang, Y.; Fang, W.; Zheng, Y.; Li, B.; Hu, Y.; Lang, C.; Huang, D.; Sun, Q.; Xiong, Y.; et al. Clinical features and treatment of COVID-19 patients in northeast Chongqing. J. Med. Virol. 2020, 92, 797–806. [Google Scholar] [CrossRef]
- Reimann, C.-M.; Gräler, M. Extraction and Quantification of Sphingosine 1-Phosphate (S1P). Bio-Protocol 2016, 6, e1817. [Google Scholar] [CrossRef]
- Andréani, P.; Gräler, M.H. Comparative quantification of sphingolipids and analogs in biological samples by high-performance liquid chromatography after chloroform extraction. Anal. Biochem. 2006, 358, 239–246. [Google Scholar] [CrossRef]
- Shaner, R.L.; Allegood, J.C.; Park, H.; Wang, E.; Kelly, S.; Haynes, C.A.; Sullards, M.C.; Merrill, A.H. Quantitative analysis of sphingolipids for lipidomics using triple quadrupole and quadrupole linear ion trap mass spectrometers. J. Lipid Res. 2009, 50, 1692–1707. [Google Scholar] [CrossRef] [Green Version]
- Fedorova, M.; Lange, M. Evaluation of lipid quantification accuracy using HILIC and RPLC MS on the example of NIST® SRM® 1950 metabolites in human plasma. Anal. Bioanal. Chem. 2020, 412, 3573–3584. [Google Scholar] [CrossRef] [Green Version]
- R Development Core Team. R a Language and Environment for Statistical Computing: Reference Index; R Foundation for Statistical Computing: Vienna, Austria, 2010. [Google Scholar]
- RStudio Team. RStudio: Integrated development environment for R. In RStudio: Integrated Development Environment for R; RStudio Team: Boston, MA, USA, 2019. [Google Scholar]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [Green Version]
- Benjaminit, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Zhao, S.; Fernald, R.D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 2005, 12, 1047–1064. [Google Scholar] [CrossRef] [Green Version]
- Hellemans, J.; Mortier, G.; de Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2008, 8, R19. [Google Scholar] [CrossRef] [Green Version]
- Epskamp, S.; Cramer, A.O.J.; Waldorp, L.J.; Schmittmann, V.D.; Borsboom, D. qgraph: Network Visualizations of Relationships in Psychometric Data. J. Stat. Softw. 2012, 48, 1–18. Available online: http://www.jstatsoft.org/ (accessed on 19 May 2023). [CrossRef] [Green Version]
- Szymańska, E.; Saccenti, E.; Smilde, A.K.; Westerhuis, J.A. Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics 2012, 8, 3–16. [Google Scholar] [CrossRef] [Green Version]
- Mühle, C.; Bilbao Canalejas, R.D.; Kornhuber, J. Sphingomyelin Synthases in Neuropsychiatric Health and Disease. Neurosignals 2019, 27, 54–76. [Google Scholar] [CrossRef]
- Cuvillier, O.; Pirianov, G.; Kleuser, B.; Vanek, P.G.; Coso, O.A.; Gutkind, J.S.; Spiegel, S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 1996, 381, 800–803. [Google Scholar] [CrossRef]
- Sassa, T.; Hirayama, T.; Kihara, A. Enzyme Activities of the Ceramide Synthases CERS2–6 Are Regulated by Phosphorylation in the C-terminal Region. J. Biol. Chem. 2016, 291, 7477–7487. [Google Scholar] [CrossRef] [Green Version]
- Coant, N.; Sakamoto, W.; Mao, C.; Hannun, Y.A. Ceramidases, roles in sphingolipid metabolism and in health and disease. Adv. Biol. Regul. 2017, 63, 122–131. [Google Scholar] [CrossRef] [Green Version]
- Prakash, H.; Upadhyay, D.; Bandapalli, O.R.; Jain, A.; Kleuser, B. Host sphingolipids: Perspective immune adjuvant for controlling SARS-CoV-2 infection for managing COVID-19 disease. Prostaglandins Other Lipid Mediat. 2021, 152, 106504. [Google Scholar] [CrossRef]
- Harvald, E.B.; Olsen, A.S.B.; Færgeman, N.J. Autophagy in the light of sphingolipid metabolism. Apoptosis 2015, 20, 658–670. [Google Scholar] [CrossRef] [Green Version]
- Ghidoni, R.; Caretti, A.; Signorelli, P. Role of Sphingolipids in the Pathobiology of Lung Inflammation. Mediat. Inflamm. 2015, 2015, 487508. [Google Scholar] [CrossRef] [Green Version]
- van Blitterswijk, W.J.; van der Luit, A.H.; Veldman, R.J.; Verheij, M.; Borst, J. Ceramide: Second messenger or modulator of membrane structure and dynamics? Biochem. J. 2003, 369, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, F.; Cecchetti, S.; Fantuzzi, L. Macrophages and Phospholipases at the Intersection between Inflammation and the Pathogenesis of HIV-1 Infection. Int. J. Mol. Sci. 2017, 18, 1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, C.J.; Smyth, M.J.; Martinet, L. Molecular mechanisms of natural killer cell activation in response to cellular stress. Cell Death Differ. 2014, 21, 5–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, A.; Guo, Y. Acid sphingomyelinase mediates human CD4+ T-cell signaling: Potential roles in T-cell responses and diseases. Cell Death Dis. 2017, 8, e2963. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Li, C.; Kadow, S.; Henry, B.D.; Steinmann, J.; Becker, K.A.; Riehle, A.; Beckmann, N.; Wilker, B.; Li, P.-L.; et al. Acid sphingomyelinase inhibition protects mice from lung edema and lethal Staphylococcus aureus sepsis. J. Mol. Med. 2015, 93, 675–689. [Google Scholar] [CrossRef] [Green Version]
- Chung, H.-Y.; Witt, C.J.; Jbeily, N.; Hurtado-Oliveros, J.; Giszas, B.; Lupp, A.; Gräler, M.H.; Bruns, T.; Stallmach, A.; Gonnert, F.A.; et al. Acid Sphingomyelinase Inhibition Prevents Development of Sepsis Sequelae in the Murine Liver. Sci. Rep. 2017, 7, 12348. [Google Scholar] [CrossRef] [Green Version]
- Wigger, D.; Schumacher, F.; Schneider-Schaulies, S.; Kleuser, B. Sphingosine 1-phosphate metabolism and insulin signaling. Cell Signal 2021, 82, 109959. [Google Scholar] [CrossRef]
- Chaurasia, B.; Summers, S.A. Ceramides in Metabolism: Key Lipotoxic Players. Annu. Rev. Physiol. 2021, 83, 303–330. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, D.X.; Tam, J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008, 369, 344–349. [Google Scholar] [CrossRef]
- Noriyuki, O.; Masafumi, S.; Kyoko, S.; Kiyoko, O.; Yoshio, M.; Kentaro, H.; Makoto, T. Both Sphingomyelin and Cholesterol in the Host Cell Membrane Are Essential for Rubella Virus Entry. J. Virol. 2017, 92, 10-1128. [Google Scholar] [CrossRef]
- Radenkovic, D.; Chawla, S.; Pirro, M.; Sahebkar, A.; Banach, M. Cholesterol in Relation to COVID-19: Should We Care about It? J. Clin. Med. 2020, 9, 1909. [Google Scholar] [CrossRef]
- Audi, A.; Soudani, N.; Dbaibo, G.; Zaraket, H. Depletion of Host and Viral Sphingomyelin Impairs Influenza Virus Infection. Front. Microbiol. 2020, 11, 612. [Google Scholar] [CrossRef]
- Abusukhun, M.; Winkler, M.S.; Pöhlmann, S.; Moerer, O.; Meissner, K.; Tampe, B.; Hofmann-Winkler, H.; Bauer, M.; Gräler, M.H.; Claus, R.A. Activation of Sphingomyelinase-Ceramide-Pathway in COVID-19 Purposes Its Inhibition for Therapeutic Strategies. Front. Immunol. 2021, 12, 784989. [Google Scholar] [CrossRef]
- Creeden, J.F.; Imami, A.S.; Eby, H.M.; Gillman, C.; Becker, K.N.; Reigle, J.; Andari, E.; Pan, Z.K.; O’Donovan, S.M.; McCullumsmith, R.E.; et al. Fluoxetine as an anti-inflammatory therapy in SARS-CoV-2 infection. Biomed. Pharmacother. 2021, 138, 111437. [Google Scholar] [CrossRef]
- Sukhatme, V.P.; Reiersen, A.M.; Vayttaden, S.J.; Sukhatme, V.V. Fluvoxamine: A Review of Its Mechanism of Action and Its Role in COVID-19. Front. Pharmacol. 2021, 12, 652688. [Google Scholar] [CrossRef]
- Whitley, R. Molnupiravir—A Step toward Orally Bioavailable Therapies for COVID-19. N. Engl. J. Med. 2021, 386, 592–593. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, L. In the age of Omicron variant: Paxlovid raises new hopes of COVID-19 recovery. J. Med. Virol. 2022, 94, 1766–1767. [Google Scholar] [CrossRef]




| Variables | Healthy Controls n = 55 | COVID-19 Patients n = 204 | COVID-19 Patients | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| Asy-to-Mild n = 36 | Moderate n = 60 | Severe n = 67 | Critical n = 41 | Convalescent n = 77 | ||||
| Demographic characteristics | ||||||||
| Age, M ± SD, and (IQR) | 35 ± 12.9 (19–69) | 55 ± 19 (20–96) | 37.5 ± 11.4 (21–67) | 49 ± 18 (24–92) | 63 ± 15.9 (30–96) | 71 ± 17.1 (20–94) | 46 ± 9.7 (30–66) | a,d,e < 0.0001; c 0.0002 f 0.0041 |
| Age < 50, n (%) | 45 (81.8) | 79 (38.7) | 28 (77.8) | 31 (51.7) | 17 (19.4) | 7 (17.1) | 42 (54.5) | a,d,e < 0.0001, c 0.0008 f 0.0014 |
| Age ≥ 50, n (%) | 10 (18.2) | 125 (61.3) | 8 (22.2) | 29 (48.3) | 54 (80.6) | 34 (82.9) | 35 (45.4) | |
| Sex, n (%) | ||||||||
| Man | 24 (43.6) | 116 (56.9) | 15 (41.7) | 36 (60) | 40 (59.7) | 25 (61) | 9 (11.7) | f < 0.0001 |
| Woman | 31 (56.4) | 88 (43.1) | 21 (58.3) | 24 (40) | 27 (40.3) | 16 (39) | 68 (88.3) | |
| BMI (kg/m3) | 25.4 ± 4.2 (15.4–34.9) | 28.4 ± 5.9 (15.8–50.3) | 27.8 ± 5.3 (15.8–43.8) | 28.3 ± 5.7 (17.4–42.1) | 28.1 ± 6.1 (20.2–47.7) | 29.4 ± 6.1 (21.7–50.3) | 29 ± 5.1 (20.7–45.5) | a 0.0002; c 0.0240 d 0.0007 e 0.0003 f 0.0041 |
| Comorbidities, n (%) | ||||||||
| Hypertension | 6 (10.9) | 90 (44.1) | 2 (5.5) | 19 (31.7) | 46 (68.6) | 23 (56.1) | 18 (23.4) | a,d,e < 0.0001; c 0.0118 |
| Cardiovascular disorder | 7 (12.7) | 21 (10.3) | 4 (11.1) | 9 (15) | 6 (8.9) | 2 (4.9) | - | |
| Diabetes mellitus | 3 (5.4) | 62 (30.4) | 3 (8.3) | 16 (32) | 29 (43.3) | 14 (34.1) | 13 (16.9) | a,d < 0.0001; c 0.0006 e 0.0004 |
| History of smoking | 6 (10.9) | 39 (19.1) | 4 (11.1) | 9 (15) | 15 (22.4) | 11 (26.8) | 2 (2.6) | |
| Neurological disorder | - | 34 (16.7) | 9 (25) | 10 (16.7) | 10 (14.9) | 5 (12.2) | 14 (18.2) | |
| Presenting symptoms, n (%) | ||||||||
| Dyspnea | - | 127 (62.2) | - | 45 (75) | 47 (70.1) | 35 (85.4) | 60 (77.9) | f 0.0157 |
| Fever | - | 64 (31.4) | 2 (5.5) | 14 (23.3) | 33 (49.2) | 15 (36.6) | 53 (68.8) | f < 0.0001 |
| Myalgia | - | 45 (22.1) | - | 7 (11.7) | 23 (34.3) | 15 (36.6) | 68 (88.3) | f < 0.0001 |
| Diarrhea | - | 52 (25.5) | 12 (33.3) | 21 (35) | 14 (20.9) | 5 (12.2) | 47 (61.1) | f < 0.0001 |
| Cough | - | 145 (71.1) | 26 (72.2) | 42 (70) | 51 (76.1) | 26 (63.4) | 53 (93) | f 0.0004 |
| Hyperactive delirium | - | 12 (5.9) | - | 5 (8.3) | - | 7 (17.1) | - | |
| Dysgeusia | - | 53 (26) | 21 (58.3) | 22 (36.7) | 8 (12) | 2 (4.9) | 62 (80.5) | f < 0.0001 |
| Anosmia | - | 58 (28.4) | 22 (61.1) | 23 (38.3) | 11 (16.4) | 2 (4.9) | 58 (75.3) | f < 0.0001 |
| Laboratory findings, M ± SD, and (IQR) | ||||||||
| Erythrocytes × 109/L | 4.7 ± 0.5 (3.6–5.8) | 4.5 ± 0.7 (2.2–5.9) | 4.8 ± 0.5 (3.9–5.8) | 4.5 ± 0.6 (3.0–5.9) | 4.3 ± 0.8 (2.2–5.8) | 4.0 ± 0.8 (2.3–5.7) | 4.6 ± 0.4 (3.7–5.4) | a 0.0076; d 0.0026; e < 0.0001 |
| Hemoglobin (g/dL) | 14.5 ± 1.5 (10.5–17.4) | 13.3 ± 2.4 (6.6–18.2) | 15 ± 1.2 (12–16.9) | 13.6 ± 2.2 (8.1–18.2) | 12.6 ± 2.3 (6.8–16.5) | 12.4 ± 2.6 (6.6–18.2) | 13.8 ± 1.4 (9.4–16.5) | a,d,e < 0.0001; c 0.0142 |
| Leukocytes × 109/L | 7.4 ± 1.8 (4.1–11.3) | 8.4 ± 4.4 (1.6–26.1) | 7.3 ± 2.3 (3.2–13.6) | 7.4 ± 2.7 (2.6–15.7) | 8.6 ± 4.1 (1.6–21.9) | 11.1 ± 6.0 (4.6–26.1) | 5.9 ± 1.8 (2.1–12.3) | e < 0.0001; f 0.0098 |
| Neutrophils × 109/L | 4.3 ± 1.3 (2.3–7.4) | 6.0 ± 4.1 (1.6–23.8) | 4.1 ± 1.7 (1.6–9.9) | 5.0 ± 2.6 (1.6–13.4) | 7.2 ± 3.5 (2.9–18.8) | 9.5 ± 5.2 (3.2–23.7) | 3.1 ± 1.3 (1.1–8.6) | a,d,e < 0.0001; f 0.0299 |
| Lymphocytes × 109/L | 2.3 ± 0.6 (1.0–3.9) | 1.3 ± 0.9 (0.1–4.3) | 2.3 ± 0.7 (1.1–4.3) | 1.5 ± 0.8 (0.3–3.8) | 1.0 ± 0.6 (0.1–2.8) | 1.0 ± 0.5 (0.2–2.2) | 2.1 ± 0.5 (1.0–3.6) | a,d,e < 0.0001; c 0.0004 |
| Neutrophil–lymphocyte ratio | 1.9 ± 0.6 (1.0–3.3) | 4.9 ± 5.6 (0.2–28.7) | 1.7 ± 0.6 (0.7–3.6) | 3.3 ± 3.1 (0.6–15.2) | 6.8 ± 4.3 (1.0–23) | 9.1 ± 6.7 (2.3–26.7) | 1.5 ± 0.7 (0.5–4.3) | a,d,e < 0.0001; c 0.0145 |
| Monocytes × 109/L | 0.5 ± 0.1 (0.3–0.9) | 0.5 ± 0.3 (0.1–1.6) | 0.5 ± 0.1 (0.2–0.9) | 0.4 ± 0.2 (0.1–1.1) | 0.5 ± 0.3 (0.1–1.3) | 0.5 ± 0.4 (0.1–1.6) | 0.4 ± 0.1 (0.2–1.0) | |
| Platelets × 109/L | 212 ± 43.8 (129–363) | 235 ± 89.5 (50–515) | 233 ± 63.1 (135–365) | 228 ± 93.8 (117–515) | 257 ± 102 (85–506) | 212 ±67 (50–370) | 213 ± 54.6 (116–386) | |
| Glycemia (mg/dL) | 89 ± 14.6 (63–146) | 114.5 ± 69 (65–409) | 87 ± 13.4 (71–127) | 101 ± 33 (65–2003) | 132 ± 78.4 (89–409) | 143 ± 81 (79–384) | 98.5± 18.6 (67–168) | a,d,e < 0.0001; c 0.0109 |
| Hospital support, n (%) | ||||||||
| Infirmary | - | 100 (49.0) | - | 34 (56.7) | 63 (94) | 3 (7.3) | - | |
| Intensive care unit | - | 44 (21.6) | - | 2 (3.3) | 4 (6.0) | 38 (92.7) | - | |
| Hospitalization data, n | ||||||||
| Days in hospital | - | 9 ± 4.1 (1–19) | 12 ± 4.9 (2–18) | 9 ± 4.0 (1–19) | 7 ± 3.2 (1–17) | 9 ± 3.8 (4–19) | - | |
| Days from symptom onset to recruitment | - | 4 ± 4.2 (1–17) | 9 ± 3.7 (2–17) | 4 ± 3.9 (1–15) | 3 ± 3.5 (1–16) | 3 ± 4.6 (1–16) | - | |
| Days recovery until recruitment | - | - | - | - | - | - | 30 ± 17.4 (15–90) | |
| Respiratory support received (%) | ||||||||
| High flow nasal cannula | - | 65 (31.9) | - | 24 (40) | 39 (58.2) | 2 (4.8) | - | |
| Oxygen masks/noninvasive | - | 35 (17.1) | - | 3 (5) | 26 (38.8) | 6 (14.6) | - | |
| Invasive ventilation | - | 33 (16.2) | - | - | 1 (1.5) | 32 (78) | - | |
| Oxygen saturation, M ± SD (IQR) | 99 ± 1.8 (90–99) | 94 ± 8.1 (54–99) | 97.5 ± 1.7 (94–99) | 96 ± 3.9 (80–99) | 91 ± 8.6 (54–99) | 89 ± 9.1 (60–96) | - | a,d,e,f < 0.0001; c 0.0008 |
| Medications, n (%) | ||||||||
| Glucocorticoid | 2 (3.6) | 125 (61.3) | 5 (13.9) | 30 (50) | 55 (82.1) | 35 (85.4) | - | a < 0.0001 |
| Azithromycin | - | 121 (59.3) | 8 (22.2) | 39 (65) | 46 (68.6) | 28 (68.3) | - | |
| Ceftriaxone | - | 93 (45.6) | - | 23 (38.3) | 46 (68.7) | 24 (58.5) | - | |
| Oseltamivir | - | 60 (29.4) | 4 (11.1) | 10 (16.7) | 34 (50.7) | 12 (29.3) | - | |
| Colchicine | - | 6 (2.9) | - | 1 (1.7) | - | 5 (12.2) | - | |
| CQ/HCQs | - | 27 (13.2) | - | 4 (6.7) | 13 (19.4) | 10 (24.4) | - | |
| Anticoagulant | - | 18 (8.8) | 1 (2.8) | 7 (11.7) | 1 (1.5) | 9 (21.9) | - | |
| Ivermectin | - | 11 (5.4) | 5 (13.9) | 6 (10) | - | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toro, D.M.; da Silva-Neto, P.V.; de Carvalho, J.C.S.; Fuzo, C.A.; Pérez, M.M.; Pimentel, V.E.; Fraga-Silva, T.F.C.; Oliveira, C.N.S.; Caruso, G.R.; Vilela, A.F.L.; et al. Plasma Sphingomyelin Disturbances: Unveiling Its Dual Role as a Crucial Immunopathological Factor and a Severity Prognostic Biomarker in COVID-19. Cells 2023, 12, 1938. https://doi.org/10.3390/cells12151938
Toro DM, da Silva-Neto PV, de Carvalho JCS, Fuzo CA, Pérez MM, Pimentel VE, Fraga-Silva TFC, Oliveira CNS, Caruso GR, Vilela AFL, et al. Plasma Sphingomyelin Disturbances: Unveiling Its Dual Role as a Crucial Immunopathological Factor and a Severity Prognostic Biomarker in COVID-19. Cells. 2023; 12(15):1938. https://doi.org/10.3390/cells12151938
Chicago/Turabian StyleToro, Diana Mota, Pedro V. da Silva-Neto, Jonatan C. S. de Carvalho, Carlos A. Fuzo, Malena M. Pérez, Vinícius E. Pimentel, Thais F. C. Fraga-Silva, Camilla N. S. Oliveira, Glaucia R. Caruso, Adriana F. L. Vilela, and et al. 2023. "Plasma Sphingomyelin Disturbances: Unveiling Its Dual Role as a Crucial Immunopathological Factor and a Severity Prognostic Biomarker in COVID-19" Cells 12, no. 15: 1938. https://doi.org/10.3390/cells12151938
APA StyleToro, D. M., da Silva-Neto, P. V., de Carvalho, J. C. S., Fuzo, C. A., Pérez, M. M., Pimentel, V. E., Fraga-Silva, T. F. C., Oliveira, C. N. S., Caruso, G. R., Vilela, A. F. L., Nobre-Azevedo, P., Defelippo-Felippe, T. V., Argolo, J. G. M., Degiovani, A. M., Ostini, F. M., Feitosa, M. R., Parra, R. S., Vilar, F. C., Gaspar, G. G., ... ImmunoCovid Consortium Group. (2023). Plasma Sphingomyelin Disturbances: Unveiling Its Dual Role as a Crucial Immunopathological Factor and a Severity Prognostic Biomarker in COVID-19. Cells, 12(15), 1938. https://doi.org/10.3390/cells12151938