Hepatocyte Sirtuin 6 Protects against Atherosclerosis and Steatohepatitis by Regulating Lipid Homeostasis
Abstract
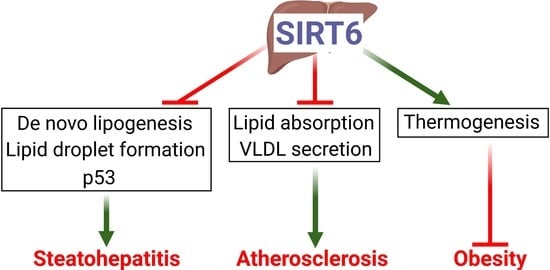
:1. Introduction
2. Materials and Methods
2.1. Mice and Diets
2.2. Adeno-Associated Virus
2.3. Real-Time PCR
2.4. Western Blot
2.5. Liver Histology and Apoptosis Assays
2.6. Hepatic Lipids and Hydroxyproline
2.7. Cell Culture and Transfection
2.8. Plasma Lipid, ALT, AST, and Lipoprotein Profile Assays
2.9. VLDL Secretion
2.10. Intestinal Fat Absorption
2.11. Intestinal Cholesterol Absorption
2.12. Bile Acid Measurement
2.13. Atherosclerotic Lesion Quantification
2.14. Body Composition and Energy Expenditure
2.15. Statistical Analysis
3. Results
3.1. Hepatocyte SIRT6 Is Required for Protection against Western Diet-Induced Steatohepatitis
3.2. Hepatic Expression of Human SIRT6 Prevents Western Diet-Induced Steatohepatitis
3.3. Hepatic SIRT6 Inhibits Genes Involved in Lipogenesis, Lipid Droplet Formation, Inflammation, and Fibrogenesis
3.4. SIRT6 Reduces Hepatic Apoptosis and Lipid Levels Partly via p53
3.5. Hepatic SIRT6 can Sufficiently Protect against the Development of Atherosclerosis in Ldlr−/− Mice
3.6. Hepatic SIRT6 is Critical for Regulating Intestinal Cholesterol and Fat Absorption and VLDL Secretion
3.7. Hepatic SIRT6 Is Required for Preventing Western Diet-Induced Obesity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kugel, S.; Mostoslavsky, R. Chromatin and beyond: The multitasking roles for SIRT6. Trends Biochem. Sci. 2014, 39, 72–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, A.R.; Ferrer, C.M.; Mostoslavsky, R. SIRT6, a Mammalian Deacylase with Multitasking Abilities. Physiol. Rev. 2020, 100, 145–169. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Xiao, C.; Wang, R.H.; Lahusen, T.; Xu, X.; Vassilopoulos, A.; Vazquez-Ortiz, G.; Jeong, W.I.; Park, O.; Ki, S.H.; et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010, 12, 224–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naiman, S.; Huynh, F.K.; Gil, R.; Glick, Y.; Shahar, Y.; Touitou, N.; Nahum, L.; Avivi, M.Y.; Roichman, A.; Kanfi, Y.; et al. SIRT6 Promotes Hepatic Beta-Oxidation via Activation of PPARalpha. Cell Rep. 2019, 29, 4127–4143.e4128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Huang, M.; Kim, H.G.; Chowdhury, K.; Gao, J.; Liu, S.; Wan, J.; Wei, L.; Dong, X.C. SIRT6 controls hepatic lipogenesis by suppressing LXR, ChREBP, and SREBP1. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166249. [Google Scholar] [CrossRef]
- Tao, R.; Xiong, X.; DePinho, R.A.; Deng, C.X.; Dong, X.C. FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J. Biol. Chem. 2013, 288, 29252–29259. [Google Scholar] [CrossRef] [Green Version]
- Tao, R.; Xiong, X.; DePinho, R.A.; Deng, C.X.; Dong, X.C. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J. Lipid Res. 2013, 54, 2745–2753. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Li, Y.; Liu, Q.; Huang, Y.; Li, R.; Wu, T.; Zhang, Z.; Zhou, J.; Huang, H.; Tang, Q.; et al. Sirt6 Alleviated Liver Fibrosis by Deacetylating Conserved Lysine 54 on Smad2 in Hepatic Stellate Cells. Hepatology 2021, 73, 1140–1157. [Google Scholar] [CrossRef]
- Zhong, X.; Huang, M.; Kim, H.G.; Zhang, Y.; Chowdhury, K.; Cai, W.; Saxena, R.; Schwabe, R.F.; Liangpunsakul, S.; Dong, X.C. SIRT6 Protects Against Liver Fibrosis by Deacetylation and Suppression of SMAD3 in Hepatic Stellate Cells. Cell. Mol. Gastroenterol. Hepatol. 2020, 10, 341–364. [Google Scholar] [CrossRef]
- Maity, S.; Muhamed, J.; Sarikhani, M.; Kumar, S.; Ahamed, F.; Spurthi, K.M.; Ravi, V.; Jain, A.; Khan, D.; Arathi, B.P.; et al. Sirtuin 6 deficiency transcriptionally up-regulates TGF-beta signaling and induces fibrosis in mice. J. Biol. Chem. 2020, 295, 415–434. [Google Scholar] [CrossRef]
- Chowdhury, K.; Huang, M.; Kim, H.G.; Dong, X.D. Sirtuin 6 protects against hepatic fibrogenesis by suppressing the YAP and TAZ function. FASEB J. 2022, 36, e22529, Erratum in FASEB J. 2022, 36, e22572. [Google Scholar] [CrossRef]
- Ka, S.O.; Bang, I.H.; Bae, E.J.; Park, B.H. Hepatocyte-specific sirtuin 6 deletion predisposes to nonalcoholic steatohepatitis by up-regulation of Bach1, an Nrf2 repressor. FASEB J. 2017, 31, 3999–4010. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Y.; Jadhav, K.; Pan, X.; Zhu, Y.; Hu, S.; Chen, S.; Chen, L.; Tang, Y.; Wang, H.H.; et al. Hepatocyte ATF3 protects against atherosclerosis by regulating HDL and bile acid metabolism. Nat. Metab. 2021, 3, 59–74. [Google Scholar] [CrossRef]
- Khalifeh-Soltani, A.; McKleroy, W.; Sakuma, S.; Cheung, Y.Y.; Tharp, K.; Qiu, Y.; Turner, S.M.; Chawla, A.; Stahl, A.; Atabai, K. Mfge8 promotes obesity by mediating the uptake of dietary fats and serum fatty acids. Nat. Med. 2014, 20, 175–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turley, S.D.; Herndon, M.W.; Dietschy, J.M. Reevaluation and application of the dual-isotope plasma ratio method for the measurement of intestinal cholesterol absorption in the hamster. J. Lipid Res. 1994, 35, 328–339. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, Y.; Hu, S.; Pan, X.; Bawa, F.C.; Wang, H.H.; Wang, D.Q.; Yin, L.; Zhang, Y. Hepatocyte miR-34a is a key regulator in the development and progression of non-alcoholic fatty liver disease. Mol. Metab. 2021, 51, 101244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ge, X.; Heemstra, L.A.; Chen, W.D.; Xu, J.; Smith, J.L.; Ma, H.; Kasim, N.; Edwards, P.A.; Novak, C.M. Loss of FXR protects against diet-induced obesity and accelerates liver carcinogenesis in ob/ob mice. Mol. Endocrinol. 2012, 26, 272–280. [Google Scholar] [CrossRef]
- Flessa, C.M.; Nasiri-Ansari, N.; Kyrou, I.; Leca, B.M.; Lianou, M.; Chatzigeorgiou, A.; Kaltsas, G.; Kassi, E.; Randeva, H.S. Genetic and Diet-Induced Animal Models for Non-Alcoholic Fatty Liver Disease (NAFLD) Research. Int. J. Mol. Sci. 2022, 23, 5791. [Google Scholar] [CrossRef]
- Schuster, S.; Cabrera, D.; Arrese, M.; Feldstein, A.E. Triggering and resolution of inflammation in NASH. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 349–364. [Google Scholar] [CrossRef]
- Capuozzo, M.; Santorsola, M.; Bocchetti, M.; Perri, F.; Cascella, M.; Granata, V.; Celotto, V.; Gualillo, O.; Cossu, A.M.; Nasti, G.; et al. p53: From Fundamental Biology to Clinical Applications in Cancer. Biology 2022, 11, 1325. [Google Scholar] [CrossRef] [PubMed]
- Derdak, Z.; Villegas, K.A.; Harb, R.; Wu, A.M.; Sousa, A.; Wands, J.R. Inhibition of p53 attenuates steatosis and liver injury in a mouse model of non-alcoholic fatty liver disease. J. Hepatol. 2013, 58, 785–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomita, K.; Teratani, T.; Suzuki, T.; Oshikawa, T.; Yokoyama, H.; Shimamura, K.; Nishiyama, K.; Mataki, N.; Irie, R.; Minamino, T.; et al. p53/p66Shc-mediated signaling contributes to the progression of non-alcoholic steatohepatitis in humans and mice. J. Hepatol. 2012, 57, 837–843. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.; Rymarchyk, S.; Zheng, S.; Cen, Y. Trichostatin A inhibits deacetylation of histone H3 and p53 by SIRT6. Arch. Biochem. Biophys. 2018, 638, 8–17. [Google Scholar] [CrossRef]
- Kanfi, Y.; Peshti, V.; Gil, R.; Naiman, S.; Nahum, L.; Levin, E.; Kronfeld-Schor, N.; Cohen, H.Y. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell 2010, 9, 162–173. [Google Scholar] [CrossRef]
- Schwarz, M.; Russell, D.W.; Dietschy, J.M.; Turley, S.D. Alternate pathways of bile acid synthesis in the cholesterol 7alpha-hydroxylase knockout mouse are not upregulated by either cholesterol or cholestyramine feeding. J. Lipid Res. 2001, 42, 1594–1603. [Google Scholar] [CrossRef]
- Li, T.; Owsley, E.; Matozel, M.; Hsu, P.; Novak, C.M.; Chiang, J.Y. Transgenic expression of cholesterol 7alpha-hydroxylase in the liver prevents high-fat diet-induced obesity and insulin resistance in mice. Hepatology 2010, 52, 678–690. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.; Zhang, Y. Hepatocyte nuclear factor 4alpha in the pathogenesis of non-alcoholic fatty liver disease. Chin. Med. J. 2022, 135, 1172–1181. [Google Scholar] [CrossRef]
- Liao, C.Y.; Song, M.J.; Gao, Y.; Mauer, A.S.; Revzin, A.; Malhi, H. Hepatocyte-Derived Lipotoxic Extracellular Vesicle Sphingosine 1-Phosphate Induces Macrophage Chemotaxis. Front. Immunol. 2018, 9, 2980. [Google Scholar] [CrossRef] [Green Version]
- Loomba, R.; Friedman, S.L.; Shulman, G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 2021, 184, 2537–2564. [Google Scholar] [CrossRef]
- Kuang, J.; Chen, L.; Tang, Q.; Zhang, J.; Li, Y.; He, J. The Role of Sirt6 in Obesity and Diabetes. Front. Physiol. 2018, 9, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Hu, S.; Pan, X.; Gopoju, R.; Cassim Bawa, F.N.; Yin, L.; Xu, Y.; Zhang, Y. Hepatocyte Sirtuin 6 Protects against Atherosclerosis and Steatohepatitis by Regulating Lipid Homeostasis. Cells 2023, 12, 2009. https://doi.org/10.3390/cells12152009
Zhu Y, Hu S, Pan X, Gopoju R, Cassim Bawa FN, Yin L, Xu Y, Zhang Y. Hepatocyte Sirtuin 6 Protects against Atherosclerosis and Steatohepatitis by Regulating Lipid Homeostasis. Cells. 2023; 12(15):2009. https://doi.org/10.3390/cells12152009
Chicago/Turabian StyleZhu, Yingdong, Shuwei Hu, Xiaoli Pan, Raja Gopoju, Fathima N. Cassim Bawa, Liya Yin, Yanyong Xu, and Yanqiao Zhang. 2023. "Hepatocyte Sirtuin 6 Protects against Atherosclerosis and Steatohepatitis by Regulating Lipid Homeostasis" Cells 12, no. 15: 2009. https://doi.org/10.3390/cells12152009
APA StyleZhu, Y., Hu, S., Pan, X., Gopoju, R., Cassim Bawa, F. N., Yin, L., Xu, Y., & Zhang, Y. (2023). Hepatocyte Sirtuin 6 Protects against Atherosclerosis and Steatohepatitis by Regulating Lipid Homeostasis. Cells, 12(15), 2009. https://doi.org/10.3390/cells12152009