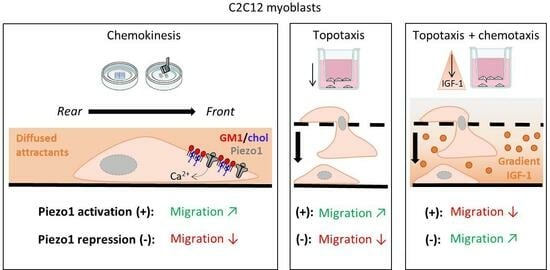
Piezo1 Is Required for Myoblast Migration and Involves Polarized Clustering in Association with Cholesterol and GM1 Ganglioside
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Chemical Treatments
2.2. Transfection of Piezo1 siRNAs or Piezo1 Fluorescent Constructs
2.3. Ibidi Chamber or Transwell Migration Assays
2.4. Calcium Live Cell Imaging
2.5. Fura2 Polarization
2.6. Focal Adhesion, Piezo1 and PMCA Immunofluorescence and Quantification
2.7. Live Cell Imaging of Plasma Membrane Cholesterol, GM1 Ganglioside, and Sphingomyelin, and the Quantification of Polarization and Distribution in Domains
2.8. Fluorescence Recovery after Photobleaching of Membrane Lipids and Piezo1
2.9. Real-Time Quantitative PCR Analysis of Piezo1
2.10. Data Presentation and Statistical Analyses
3. Results
3.1. Piezo1 Silencing in Myoblasts and Piezo1 Knockout in HEK Cells Decrease Migration in Ibidi and Transwell Chambers, While Piezo1 Activation in Myoblasts Acts in an Opposite Manner
3.2. Piezo1 Activation Increases Calcium Levels, Particularly at the Front of Migrating Myoblasts
3.3. Piezo1 Gathers in Clusters and Polarizes at the Migration Front of Myoblasts, but Its Activation Does Not Modulate the Focal Adhesion Number or Distribution
3.4. Cholesterol Depletion Impairs Piezo1 Clustering, and Reversibly Abrogates Piezo1 Polarization upon Myoblast Migration
3.5. Piezo1 Clusters Partially Associate with Cholesterol- and GM1-Enriched Domains in Migrating Myoblasts
3.6. The Mobile Fraction of GM1 Associated with the Domains Is Lower than in the Bulk Membrane at the Front of the Migrating Myoblasts Activated for Piezo1
3.7. GM1-Containing Domains Are More Abundant upon Piezo1 Activation in Myoblasts and Are Impaired upon Piezo1 Silencing or Deletion in Myoblasts and HEK Cells
3.8. The Differential Mobile Fraction of Piezo1 between the Clusters and the Bulk Membrane Is Abrogated by Cholesterol Depletion in Myoblasts
3.9. Cholesterol Depletion Abrogates Both Piezo1 and PMCA Polarization and Spatial Dissociation upon Myoblast Migration
4. Discussion
4.1. Piezo1 Differential Implication in Cell Migration Based on Migration Mode
4.2. Piezo1 and Ca2+ Polarization at the Migration Front upon Activation
4.3. Piezo1 Association with Specific Lipid Domains at the Migration Front
4.4. Lipid-Dependent Spatial Dissociation of Piezo1 and PMCA at the Front
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Lai, A.; Cox, C.D.; Chandra Sekar, N.; Thurgood, P.; Jaworowski, A.; Peter, K.; Baratchi, S. Mechanosensing by Piezo1 and Its Implications for Physiology and Various Pathologies. Biol. Rev. 2022, 97, 604–614. [Google Scholar] [CrossRef] [PubMed]
- De Felice, D.; Alaimo, A. Mechanosensitive Piezo Channels in Cancer: Focus on Altered Calcium Signaling in Cancer Cells and in Tumor Progression. Cancers 2020, 12, 1780. [Google Scholar] [CrossRef] [PubMed]
- Tsai, F.-C.; Kuo, G.-H.; Chang, S.-W.; Tsai, P.-J. Ca2+ Signaling in Cytoskeletal Reorganization, Cell Migration, and Cancer Metastasis. BioMed Res. Int. 2015, 2015, e409245. [Google Scholar] [CrossRef]
- Zhu, T.; Guo, J.; Wu, Y.; Lei, T.; Zhu, J.; Chen, H.; Kala, S.; Wong, K.F.; Cheung, C.P.; Huang, X.; et al. The Mechanosensitive Ion Channel Piezo1 Modulates the Migration and Immune Response of Microglia. iScience 2023, 26, 105993. [Google Scholar] [CrossRef]
- Botello-Smith, W.M.; Jiang, W.; Zhang, H.; Ozkan, A.D.; Lin, Y.-C.; Pham, C.N.; Lacroix, J.J.; Luo, Y. A Mechanism for the Activation of the Mechanosensitive Piezo1 Channel by the Small Molecule Yoda1. Nat. Commun. 2019, 10, 4503. [Google Scholar] [CrossRef]
- Jäntti, H.; Sitnikova, V.; Ishchenko, Y.; Shakirzyanova, A.; Giudice, L.; Ugidos, I.F.; Gómez-Budia, M.; Korvenlaita, N.; Ohtonen, S.; Belaya, I.; et al. Microglial Amyloid Beta Clearance Is Driven by PIEZO1 Channels. J. Neuroinflammation 2022, 19, 147. [Google Scholar] [CrossRef]
- Holt, J.R.; Zeng, W.-Z.; Evans, E.L.; Woo, S.-H.; Ma, S.; Abuwarda, H.; Loud, M.; Patapoutian, A.; Pathak, M.M. Spatiotemporal Dynamics of PIEZO1 Localization Controls Keratinocyte Migration during Wound Healing. eLife 2021, 10, e65415. [Google Scholar] [CrossRef]
- Li, J.; Hou, B.; Tumova, S.; Muraki, K.; Bruns, A.; Ludlow, M.J.; Sedo, A.; Hyman, A.J.; McKeown, L.; Young, R.S.; et al. Piezo1 Integration of Vascular Architecture with Physiological Force. Nature 2014, 515, 279–282. [Google Scholar] [CrossRef]
- Yu, Y.; Wu, X.; Liu, S.; Zhao, H.; Li, B.; Zhao, H.; Feng, X. Piezo1 Regulates Migration and Invasion of Breast Cancer Cells via Modulating Cell Mechanobiological Properties. Acta Biochim. Biophys. Sin. 2021, 53, 10–18. [Google Scholar] [CrossRef] [PubMed]
- SenGupta, S.; Parent, C.A.; Bear, J.E. The Principles of Directed Cell Migration. Nat. Rev. Mol. Cell Biol. 2021, 22, 529–547. [Google Scholar] [CrossRef]
- Vasileva, V.; Chubinskiy-Nadezhdin, V. Regulation of PIEZO1 Channels by Lipids and the Structural Components of Extracellular Matrix/Cell Cytoskeleton. J. Cell. Physiol. 2023, 238, 918–930. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhou, H.; Chi, S.; Wang, Y.; Wang, J.; Geng, J.; Wu, K.; Liu, W.; Zhang, T.; Dong, M.-Q.; et al. Structure and Mechanogating Mechanism of the Piezo1 Channel. Nature 2018, 554, 487–492. [Google Scholar] [CrossRef]
- Saotome, K.; Murthy, S.E.; Kefauver, J.M.; Whitwam, T.; Patapoutian, A.; Ward, A.B. Structure of the Mechanically Activated Ion Channel Piezo1. Nature 2018, 554, 481–486. [Google Scholar] [CrossRef]
- Wang, Y.; Chi, S.; Guo, H.; Li, G.; Wang, L.; Zhao, Q.; Rao, Y.; Zu, L.; He, W.; Xiao, B. A Lever-like Transduction Pathway for Long-Distance Chemical- and Mechano-Gating of the Mechanosensitive Piezo1 Channel. Nat. Commun. 2018, 9, 1300. [Google Scholar] [CrossRef]
- Mulhall, E.M.; Gharpure, A.; Lee, R.M.; Dubin, A.E.; Aaron, J.S.; Marshall, K.L.; Spencer, K.R.; Reiche, M.A.; Henderson, S.C.; Chew, T.-L.; et al. Direct Observation of the Conformational States of PIEZO1. Nature 2023, 620, 1117–1125. [Google Scholar] [CrossRef]
- Guo, Y.R.; MacKinnon, R. Structure-Based Membrane Dome Mechanism for Piezo Mechanosensitivity. eLife 2017, 6, e33660. [Google Scholar] [CrossRef]
- Yang, S.; Miao, X.; Arnold, S.; Li, B.; Ly, A.T.; Wang, H.; Wang, M.; Guo, X.; Pathak, M.M.; Zhao, W.; et al. Membrane Curvature Governs the Distribution of Piezo1 in Live Cells. Nat. Commun. 2022, 13, 7467. [Google Scholar] [CrossRef]
- Wang, S.; Wang, B.; Shi, Y.; Möller, T.; Stegmeyer, R.I.; Strilic, B.; Li, T.; Yuan, Z.; Wang, C.; Wettschureck, N.; et al. Mechanosensation by Endothelial PIEZO1 Is Required for Leukocyte Diapedesis. Blood 2022, 140, 171–183. [Google Scholar] [CrossRef]
- Cox, C.D.; Bae, C.; Ziegler, L.; Hartley, S.; Nikolova-Krstevski, V.; Rohde, P.R.; Ng, C.-A.; Sachs, F.; Gottlieb, P.A.; Martinac, B. Removal of the Mechanoprotective Influence of the Cytoskeleton Reveals PIEZO1 Is Gated by Bilayer Tension. Nat. Commun. 2016, 7, 10366. [Google Scholar] [CrossRef]
- Conrard, L.; Tyteca, D. Regulation of Membrane Calcium Transport Proteins by the Surrounding Lipid Environment. Biomolecules 2019, 9, 513. [Google Scholar] [CrossRef]
- Ridone, P.; Pandzic, E.; Vassalli, M.; Cox, C.D.; Macmillan, A.; Gottlieb, P.A.; Martinac, B. Disruption of Membrane Cholesterol Organization Impairs the Activity of PIEZO1 Channel Clusters. J. Gen. Physiol. 2020, 152, e201912515. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Ferrari, G.; Tedesco, F.S. Cellular Dynamics of Myogenic Cell Migration: Molecular Mechanisms and Implications for Skeletal Muscle Cell Therapies. EMBO Mol. Med. 2020, 12, e12357. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, K.; Takano, K.; Suetsugu, S.; Kurisu, S.; Yamazaki, D.; Miki, H.; Takenawa, T.; Endo, T. N-WASP and WAVE2 Acting Downstream of Phosphatidylinositol 3-Kinase Are Required for Myogenic Cell Migration Induced by Hepatocyte Growth Factor. J. Biol. Chem. 2004, 279, 54862–54871. [Google Scholar] [CrossRef] [PubMed]
- Méhes, E.; Mones, E.; Németh, V.; Vicsek, T. Collective Motion of Cells Mediates Segregation and Pattern Formation in Co-Cultures. PLoS ONE 2012, 7, e31711. [Google Scholar] [CrossRef] [PubMed]
- Ortuste Quiroga, H.P.; Ganassi, M.; Yokoyama, S.; Nakamura, K.; Yamashita, T.; Raimbach, D.; Hagiwara, A.; Harrington, O.; Breach-Teji, J.; Asakura, A.; et al. Fine-Tuning of Piezo1 Expression and Activity Ensures Efficient Myoblast Fusion during Skeletal Myogenesis. Cells 2022, 11, 393. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, M.; Hara, Y.; Okuda, M.; Itoh, K.; Nishioka, R.; Shiomi, A.; Nagao, K.; Mori, M.; Mori, Y.; Ikenouchi, J.; et al. Cell Surface Flip-Flop of Phosphatidylserine Is Critical for PIEZO1-Mediated Myotube Formation. Nat. Commun. 2018, 9, 2049. [Google Scholar] [CrossRef] [PubMed]
- Vanderroost, J.; Avalosse, N.; Mohammed, D.; Hoffmann, D.; Henriet, P.; Pierreux, C.E.; Alsteens, D.; Tyteca, D. Cholesterol and Sphingomyelin Polarize at the Leading Edge of Migrating Myoblasts and Involve Their Clustering in Submicrometric Domains. Biomolecules 2023, 13, 319. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, V.; Mathur, J.; Mao, R.; Bayrak-Toydemir, P.; Procter, M.; Cahalan, S.M.; Kim, H.J.; Bandell, M.; Longo, N.; Day, R.W.; et al. Impaired PIEZO1 Function in Patients with a Novel Autosomal Recessive Congenital Lymphatic Dysplasia. Nat. Commun. 2015, 6, 8329. [Google Scholar] [CrossRef]
- Jackson, M.F.; Hoversten, K.E.; Powers, J.M.; Trobridge, G.D.; Rodgers, B.D. Genetic Manipulation of Myoblasts and a Novel Primary Myosatellite Cell Culture System: Comparing and Optimizing Approaches. FEBS J. 2013, 280, 827–839. [Google Scholar] [CrossRef]
- Ly, P.T.; Xu, S.; Wirawan, M.; Luo, D.; Roca, X. ZAP Isoforms Regulate Unfolded Protein Response and Epithelial- Mesenchymal Transition. Proc. Natl. Acad. Sci. USA 2022, 119, e2121453119. [Google Scholar] [CrossRef]
- Tinning, P.W.; Franssen, A.J.P.M.; Hridi, S.U.; Bushell, T.J.; Mcconnell, G. A 340/380 Nm Light-Emitting Diode Illuminator for Fura-2 AM Ratiometric Ca2+ Imaging of Live Cells with Better than 5 nM Precision. J. Microsc. 2018, 269, 212–220. [Google Scholar] [CrossRef] [PubMed]
- McHugh, B.J.; Buttery, R.; Lad, Y.; Banks, S.; Haslett, C.; Sethi, T. Integrin Activation by Fam38A Uses a Novel Mechanism of R-Ras Targeting to the Endoplasmic Reticulum. J. Cell Sci. 2010, 123, 51–61. [Google Scholar] [CrossRef] [PubMed]
- McHugh, B.J.; Murdoch, A.; Haslett, C.; Sethi, T. Loss of the Integrin-Activating Transmembrane Protein Fam38A (Piezo1) Promotes a Switch to a Reduced Integrin-Dependent Mode of Cell Migration. PLoS ONE 2012, 7, e40346. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Tijore, A.; Cheng, D.; Li, J.V.; Hariharan, A.; Martinac, B.; Tran Van Nhieu, G.; Cox, C.D.; Sheetz, M. Force- and Cell State-Dependent Recruitment of Piezo1 Drives Focal Adhesion Dynamics and Calcium Entry. Sci. Adv. 2022, 8, eabo1461. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.; Thurgood, P.; Cox, C.D.; Chheang, C.; Peter, K.; Jaworowski, A.; Khoshmanesh, K.; Baratchi, S. Piezo1 Response to Shear Stress Is Controlled by the Components of the Extracellular Matrix. ACS Appl. Mater. Interfaces 2022, 14, 40559–40568. [Google Scholar] [CrossRef] [PubMed]
- Tsai, F.-C.; Seki, A.; Yang, H.W.; Hayer, A.; Carrasco, S.; Malmersjö, S.; Meyer, T. A Polarized Ca2+, Diacylglycerol, and STIM1 Signaling System Regulates Directed Cell Migration. Nat. Cell Biol. 2014, 16, 133–144. [Google Scholar] [CrossRef]
- Wilkinson, P.C. Random Locomotion; Chemotaxis and Chemokinesis. A Guide to Terms Defining Cell Locomotion. Immunol. Today 1985, 6, 273–278. [Google Scholar] [CrossRef]
- Canales Coutiño, B.; Mayor, R. Reprint of: Mechanosensitive Ion Channels in Cell Migration. Cells Dev. 2021, 168, 203730. [Google Scholar] [CrossRef]
- Seetharaman, S.; Etienne-Manneville, S. Cytoskeletal Crosstalk in Cell Migration. Trends Cell Biol. 2020, 30, 720–735. [Google Scholar] [CrossRef]
- Trepat, X.; Wasserman, M.R.; Angelini, T.E.; Millet, E.; Weitz, D.A.; Butler, J.P.; Fredberg, J.J. Physical Forces during Collective Cell Migration. Nat. Phys. 2009, 5, 426–430. [Google Scholar] [CrossRef]
- Goetsch, K.P.; Kallmeyer, K.; Niesler, C.U. Decorin Modulates Collagen I-Stimulated, but Not Fibronectin-Stimulated, Migration of C2C12 Myoblasts. Matrix Biol. 2011, 30, 109–117. [Google Scholar] [CrossRef]
- Jetta, D.; Shireen, T.; Hua, S.Z. Epithelial Cells Sense Local Stiffness via Piezo1 Mediated Cytoskeletal Reorganization. Front. Cell Dev. Biol. 2023, 11, 1198109. [Google Scholar] [CrossRef] [PubMed]
- Gkretsi, V.; Stylianopoulos, T. Cell Adhesion and Matrix Stiffness: Coordinating Cancer Cell Invasion and Metastasis. Front. Oncol. 2018, 8, 145. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-J.; Le Berre, M.; Lautenschlaeger, F.; Maiuri, P.; Callan-Jones, A.; Heuzé, M.; Takaki, T.; Voituriez, R.; Piel, M. Confinement and Low Adhesion Induce Fast Amoeboid Migration of Slow Mesenchymal Cells. Cell 2015, 160, 659–672. [Google Scholar] [CrossRef] [PubMed]
- Otto, A.; Collins-Hooper, H.; Patel, A.; Dash, P.R.; Patel, K. Adult Skeletal Muscle Stem Cell Migration Is Mediated by a Blebbing/Amoeboid Mechanism. Rejuvenation Res. 2011, 14, 249–260. [Google Scholar] [CrossRef]
- Hung, W.-C.; Yang, J.R.; Yankaskas, C.L.; Wong, B.S.; Wu, P.-H.; Pardo-Pastor, C.; Serra, S.A.; Chiang, M.-J.; Gu, Z.; Wirtz, D.; et al. Confinement Sensing and Signal Optimization via Piezo1/PKA and Myosin II Pathways. Cell Rep. 2016, 15, 1430–1441. [Google Scholar] [CrossRef]
- Louis, M.; Zanou, N.; Schoor, M.V.; Gailly, P. TRPC1 Regulates Skeletal Myoblast Migration and Differentiation. J. Cell Sci. 2008, 121, 3951–3959. [Google Scholar] [CrossRef]
- Kurth, F.; Franco-Obregón, A.; Casarosa, M.; Küster, S.K.; Wuertz-Kozak, K.; Dittrich, P.S. Transient Receptor Potential Vanilloid 2-Mediated Shear-Stress Responses in C2C12 Myoblasts Are Regulated by Serum and Extracellular Matrix. FASEB J. 2015, 29, 4726–4737. [Google Scholar] [CrossRef]
- Zagranichnaya, T.K.; Wu, X.; Villereal, M.L. Endogenous TRPC1, TRPC3, and TRPC7 Proteins Combine to Form Native Store-Operated Channels in HEK-293 Cells. J. Biol. Chem. 2005, 280, 29559–29569. [Google Scholar] [CrossRef]
- Mrkonjić, S.; Garcia-Elias, A.; Pardo-Pastor, C.; Bazellières, E.; Trepat, X.; Vriens, J.; Ghosh, D.; Voets, T.; Vicente, R.; Valverde, M.A. TRPV4 Participates in the Establishment of Trailing Adhesions and Directional Persistence of Migrating Cells. Pflugers Arch.-Eur. J. Physiol. 2015, 467, 2107–2119. [Google Scholar] [CrossRef]
- Remorino, A.; Beco, S.D.; Cayrac, F.; Federico, F.D.; Cornilleau, G.; Gautreau, A.; Parrini, M.C.; Masson, J.-B.; Dahan, M.; Coppey, M. Gradients of Rac1 Nanoclusters Support Spatial Patterns of Rac1 Signaling. Cell Rep. 2017, 21, 1922–1935. [Google Scholar] [CrossRef]
- Llense, F.; Etienne-Manneville, S. Front-to-Rear Polarity in Migrating Cells. In Cell Polarity 1: Biological Role and Basic Mechanisms; Ebnet, K., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 115–146. ISBN 978-3-319-14463-4. [Google Scholar]
- Vaz, R.; Martins, G.; Thorsteinsdóttir, S.; Rodrigues, G. Fibronectin Promotes Migration, Alignment and Fusion in an in Vitro Myoblast Cell Model. Cell Tissue Res. 2012, 348, 569–578. [Google Scholar] [CrossRef]
- Du, W.; Hong, S.; Scapin, G.; Goulard, M.; Shah, D.I. Directed Collective Cell Migration Using Three-Dimensional Bioprinted Micropatterns on Thermoresponsive Surfaces for Myotube Formation. ACS Biomater. Sci. Eng. 2019, 5, 3935–3943. [Google Scholar] [CrossRef]
- Liao, J.; Lu, W.; Chen, Y.; Duan, X.; Zhang, C.; Luo, X.; Lin, Z.; Chen, J.; Liu, S.; Yan, H.; et al. Upregulation of Piezo1 (Piezo Type Mechanosensitive Ion Channel Component 1) Enhances the Intracellular Free Calcium in Pulmonary Arterial Smooth Muscle Cells From Idiopathic Pulmonary Arterial Hypertension Patients. Hypertension 2021, 77, 1974–1989. [Google Scholar] [CrossRef]
- Brundage, R.A.; Fogarty, K.E.; Tuft, R.A.; Fay, F.S. Calcium Gradients Underlying Polarization and Chemotaxis of Eosinophils. Science 1991, 254, 703–706. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Wang, X.; Chen, M.; Ouyang, K.; Song, L.-S.; Cheng, H. Calcium Flickers Steer Cell Migration. Nature 2009, 457, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.H.; Falke, J.J. Ca2+ Influx Is an Essential Component of the Positive-Feedback Loop That Maintains Leading-Edge Structure and Activity in Macrophages. Proc. Natl. Acad. Sci. USA 2007, 104, 16176–16181. [Google Scholar] [CrossRef] [PubMed]
- Ellefsen, K.L.; Holt, J.R.; Chang, A.C.; Nourse, J.L.; Arulmoli, J.; Mekhdjian, A.H.; Abuwarda, H.; Tombola, F.; Flanagan, L.A.; Dunn, A.R.; et al. Myosin-II Mediated Traction Forces Evoke Localized Piezo1-Dependent Ca2+ Flickers. Commun. Biol. 2019, 2, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Buyan, A.; Cox, C.D.; Barnoud, J.; Li, J.; Chan, H.S.M.; Martinac, B.; Marrink, S.J.; Corry, B. Piezo1 Forms Specific, Functionally Important Interactions with Phosphoinositides and Cholesterol. Biophys. J. 2020, 119, 1683–1697. [Google Scholar] [CrossRef]
- Hulce, J.J.; Cognetta, A.B.; Niphakis, M.J.; Tully, S.E.; Cravatt, B.F. Proteome-Wide Mapping of Cholesterol-Interacting Proteins in Mammalian Cells. Nat. Methods 2013, 10, 259–264. [Google Scholar] [CrossRef]
- Pontes, B.; Monzo, P.; Gole, L.; Le Roux, A.-L.; Kosmalska, A.J.; Tam, Z.Y.; Luo, W.; Kan, S.; Viasnoff, V.; Roca-Cusachs, P.; et al. Membrane Tension Controls Adhesion Positioning at the Leading Edge of Cells. J. Cell Biol. 2017, 216, 2959–2977. [Google Scholar] [CrossRef]
- Chakraborty, S.; Doktorova, M.; Molugu, T.R.; Heberle, F.A.; Scott, H.L.; Dzikovski, B.; Nagao, M.; Stingaciu, L.-R.; Standaert, R.F.; Barrera, F.N.; et al. How Cholesterol Stiffens Unsaturated Lipid Membranes. Proc. Natl. Acad. Sci. USA 2020, 117, 21896–21905. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.D.; Zhang, Y.; Zhou, Z.; Walz, T.; Martinac, B. Cyclodextrins Increase Membrane Tension and Are Universal Activators of Mechanosensitive Channels. Proc. Natl. Acad. Sci. USA 2021, 118, e2104820118. [Google Scholar] [CrossRef] [PubMed]
- Hirata, Y.; Cai, R.; Volchuk, A.; Steinberg, B.E.; Saito, Y.; Matsuzawa, A.; Grinstein, S.; Freeman, S.A. Lipid Peroxidation Increases Membrane Tension, Piezo1 Gating, and Cation Permeability to Execute Ferroptosis. Curr. Biol. 2023, 33, 1282–1294.e5. [Google Scholar] [CrossRef]
- Mim, C.; Unger, V.M. Membrane Curvature and Its Generation by BAR Proteins. Trends Biochem. Sci. 2012, 37, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Leonard, C.; Conrard, L.; Guthmann, M.; Pollet, H.; Carquin, M.; Vermylen, C.; Gailly, P.; Smissen, P.V.D.; Mingeot-Leclercq, M.P.; Tyteca, D. Contribution of Plasma Membrane Lipid Domains to Red Blood Cell (Re)Shaping. Sci. Rep. 2017, 7, 4264. [Google Scholar] [CrossRef] [PubMed]
- Dumitru, A.C.; Stommen, A.; Koehler, M.; Cloos, A.S.; Yang, J.; Leclercqz, A.; Tyteca, D.; Alsteens, D. Probing PIEZO1 Localization upon Activation Using High-Resolution Atomic Force and Confocal Microscopy. Nano Lett. 2021, 21, 4950–4958. [Google Scholar] [CrossRef]
- Conrard, L.; Stommen, A.; Cloos, A.-S.; Steinkühler, J.; Dimova, R.; Pollet, H.; Tyteca, D. Spatial Relationship and Functional Relevance of Three Lipid Domain Populations at the Erythrocyte Surface. Cell. Physiol. Biochem. 2018, 51, 1544–1565. [Google Scholar] [CrossRef]
- Pang, Y.; Zhu, H.; Wu, P.; Chen, J. The Characterization of Plasma Membrane Ca2+-ATPase in Rich Sphingomyelin–Cholesterol Domains. FEBS Lett. 2005, 579, 2397–2403. [Google Scholar] [CrossRef]
- Tang, D.; Dean, W.L.; Borchman, D.; Paterson, C.A. The Influence of Membrane Lipid Structure on Plasma Membrane Ca2+-ATPase Activity. Cell Calcium 2006, 39, 209–216. [Google Scholar] [CrossRef]
- Jiang, L.; Fernandes, D.; Mehta, N.; Bean, J.L.; Michaelis, M.L.; Zaidi, A. Partitioning of the Plasma Membrane Ca2+-ATPase into Lipid Rafts in Primary Neurons: Effects of Cholesterol Depletion. J. Neurochem. 2007, 102, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Subczynski, W.K.; Pasenkiewicz-Gierula, M.; Widomska, J.; Mainali, L.; Raguz, M. High Cholesterol/Low Cholesterol: Effects in Biological Membranes Review. Cell Biochem. Biophys. 2017, 75, 369–385. [Google Scholar] [CrossRef] [PubMed]
- Bruce, J.I.E. Metabolic Regulation of the PMCA: Role in Cell Death and Survival. Cell Calcium 2018, 69, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Sarmento, M.J.; Coutinho, A.; Fedorov, A.; Prieto, M.; Fernandes, F. Ca2+ Induces PI(4,5)P2 Clusters on Lipid Bilayers at Physiological PI(4,5)P2 and Ca2+ Concentrations. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2014, 1838, 822–830. [Google Scholar] [CrossRef]
- Abe, M.; Makino, A.; Hullin-Matsuda, F.; Kamijo, K.; Ohno-Iwashita, Y.; Hanada, K.; Mizuno, H.; Miyawaki, A.; Kobayashi, T. A Role for Sphingomyelin-Rich Lipid Domains in the Accumulation of Phosphatidylinositol-4,5-Bisphosphate to the Cleavage Furrow during Cytokinesis. Mol. Cell. Biol. 2012, 32, 1396–1407. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanderroost, J.; Parpaite, T.; Avalosse, N.; Henriet, P.; Pierreux, C.E.; Lorent, J.H.; Gailly, P.; Tyteca, D. Piezo1 Is Required for Myoblast Migration and Involves Polarized Clustering in Association with Cholesterol and GM1 Ganglioside. Cells 2023, 12, 2784. https://doi.org/10.3390/cells12242784
Vanderroost J, Parpaite T, Avalosse N, Henriet P, Pierreux CE, Lorent JH, Gailly P, Tyteca D. Piezo1 Is Required for Myoblast Migration and Involves Polarized Clustering in Association with Cholesterol and GM1 Ganglioside. Cells. 2023; 12(24):2784. https://doi.org/10.3390/cells12242784
Chicago/Turabian StyleVanderroost, Juliette, Thibaud Parpaite, Noémie Avalosse, Patrick Henriet, Christophe E. Pierreux, Joseph H. Lorent, Philippe Gailly, and Donatienne Tyteca. 2023. "Piezo1 Is Required for Myoblast Migration and Involves Polarized Clustering in Association with Cholesterol and GM1 Ganglioside" Cells 12, no. 24: 2784. https://doi.org/10.3390/cells12242784
APA StyleVanderroost, J., Parpaite, T., Avalosse, N., Henriet, P., Pierreux, C. E., Lorent, J. H., Gailly, P., & Tyteca, D. (2023). Piezo1 Is Required for Myoblast Migration and Involves Polarized Clustering in Association with Cholesterol and GM1 Ganglioside. Cells, 12(24), 2784. https://doi.org/10.3390/cells12242784