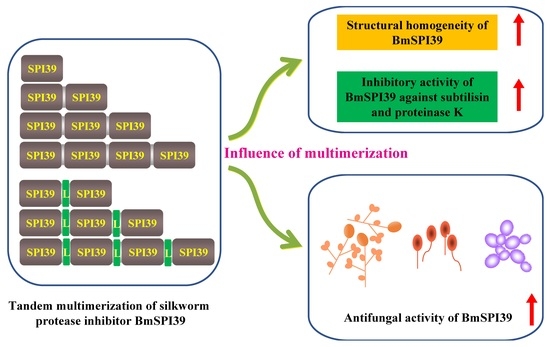
Tandem Multimerization Can Enhance the Structural Homogeneity and Antifungal Activity of the Silkworm Protease Inhibitor BmSPI39
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungi and Reagents
2.2. Vector Construction of the Basic Units
2.3. Expression Vector Construct of BmSPI39 Tandem Multimers
2.4. Protein Expression and Purification
2.5. In-Gel Activity Staining of Protease Inhibitor
2.6. Protease Inhibition Assays
2.7. Assays of Conidial Germination of B. bassiana
2.8. Fungal Growth Inhibition Assay
2.9. Statistical Analysis
3. Results
3.1. Design and Construction of Expression Vector of BmSPI39 Tandem Multimers
3.2. Protein Expression and Purification of BmSPI39 Tandem Multimers
3.3. Activity and Structural Homogeneity Analysis of BmSPI39 Tandem Multimers
3.4. Comparison of Inhibitory Capacity of BmSPI39 Tandem Multimers against Microbial Protease
3.5. Evaluation of Inhibitory Ability of BmSPI39 Tandem Multimers on Conidial Germination of Silkworm Pathogenic Fungi B. bassiana
3.6. Evaluation of the Inhibitory Effects of BmSPI39 Tandem Multimers on the Growth of Single-Celled Fungus Saccharomyces cerevisiae and Opportunistic Human Pathogen Candida albicans
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xia, Q.; Zhou, Z.; Lu, C.; Cheng, D.; Dai, F.; Li, B.; Zhao, P.; Zha, X.; Cheng, T.; Chai, C.; et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 2004, 306, 1937–1940. [Google Scholar]
- Xia, Q.; Cheng, D.; Duan, J.; Wang, G.; Cheng, T.; Zha, X.; Liu, C.; Zhao, P.; Dai, F.; Zhang, Z.; et al. Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm, Bombyx mori. Genome Biol. 2007, 8, R162. [Google Scholar] [CrossRef] [Green Version]
- Duan, J.; Li, R.; Cheng, D.; Fan, W.; Zha, X.; Cheng, T.; Wu, Y.; Wang, J.; Mita, K.; Xiang, Z.; et al. SilkDB v2.0: A platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 2010, 38, D453–D456. [Google Scholar] [CrossRef] [Green Version]
- Xia, Q.; Li, S.; Feng, Q. Advances in silkworm studies accelerated by the genome sequencing of Bombyx mori. Annu. Rev. Entomol. 2014, 59, 513–536. [Google Scholar] [CrossRef]
- Xiong, Q.; Xie, Y.P.; Zhu, Y.M.; Xue, J.L.; Li, J.; Fan, R.J. Morphological and ultrastructural characterization of Carposina sasakii larvae (Lepidoptera: Carposinidae) infected by Beauveria bassiana (Ascomycota: Hypocreales: Clavicipitaceae). Micron 2013, 44, 303–311. [Google Scholar] [CrossRef]
- Charnley, A.K. Physiological Aaspects of Destructive Pathogenesis in Insects by Fungi: A Speculative Review; Cambridge University Press: London, UK, 1984. [Google Scholar]
- Goettel, M.S.; Leger, R.J.S.; Rizzo, N.W.; Staples, R.C.; Roberts, D.W. Ultrastructural Localization of a Cuticle-degrading Protease Produced by the Entomopathogenic Fungus Metarhizium anisopliae during Penetration of Host (Manduca sexta) Cuticle. J. Gen. Microbiol. 1989, 135, 2233–2239. [Google Scholar]
- St Leger, R.J. The role of cuticle-degrading proteases in fungal pathogenesis of insects. Can. J. Bot 1995, 73, S1119–S1125. [Google Scholar] [CrossRef]
- Charnley, A.K. Fungal Pathogens of Insects: Cuticle Degrading Enzymes and Toxin. Adv. Bot. Res. 2003, 40, 241–321. [Google Scholar]
- St Leger, R.J.; Joshi, L.; Bidochka, M.J.; Roberts, D.W. Construction of an improved mycoinsecticide overexpressing a toxic protease. Proc. Natl. Acad. Sci. USA 1996, 93, 6349–6354. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.-J.; Feng, M.-G.; Fan, Y.-H.; Luo, Z.-B.; Yang, X.-Y.; Wu, D.; Pei, Y. A cuticle-degrading protease (CDEP-1) of Beauveria bassiana enhances virulence. Biocontrol. Sci. Technol. 2008, 18, 543–555. [Google Scholar] [CrossRef]
- Yang, J.; Zhao, X.; Liang, L.; Xia, Z.; Lei, L.; Niu, X.; Zou, C.; Zhang, K.Q. Overexpression of a cuticle-degrading protease Ver112 increases the nematicidal activity of Paecilomyces lilacinus. Appl. Microbiol. Biotechnol. 2011, 89, 1895–1903. [Google Scholar] [CrossRef]
- Kanost, M.R. Serine proteinase inhibitors in arthropod immunity. Dev. Comp. Immunol. 1999, 23, 291–301. [Google Scholar] [CrossRef]
- Fullaondo, A.; Garcia-Sanchez, S.; Sanz-Parra, A.; Recio, E.; Lee, S.Y.; Gubb, D. Spn1 regulates the GNBP3-dependent Toll signaling pathway in Drosophila melanogaster. Mol. Cell Biol. 2011, 31, 2960–2972. [Google Scholar] [CrossRef] [Green Version]
- Cerenius, L.; Kawabata, S.; Lee, B.L.; Nonaka, M.; Soderhall, K. Proteolytic cascades and their involvement in invertebrate immunity. Trends Biochem. Sci. 2010, 35, 575–583. [Google Scholar] [CrossRef]
- Zhao, P.; Dong, Z.; Duan, J.; Wang, G.; Wang, L.; Li, Y.; Xiang, Z.; Xia, Q. Genome-wide identification and immune response analysis of serine protease inhibitor genes in the silkworm, Bombyx mori. PLoS ONE 2012, 7, e31168. [Google Scholar] [CrossRef]
- Li, Y.; Lu, Q.; Yang, X.; Zhang, J.; Luo, Z.; Xia, Q.; Zhao, P. Expression of the fungal-resistance factor BmSPI39 in Bombyx mori in response to Beauveria bassiana invasion. Acta Entomol. Sin. 2021, 64, 61–69. [Google Scholar]
- Li, Y.; Zhao, P.; Liu, S.; Dong, Z.; Chen, J.; Xiang, Z.; Xia, Q. A novel protease inhibitor in Bombyx mori is involved in defense against Beauveria bassiana. Insect Biochem. Mol. Biol. 2012, 42, 766–775. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, P.; Liu, H.; Guo, X.; He, H.; Zhu, R.; Xiang, Z.; Xia, Q. TIL-type protease inhibitors may be used as targeted resistance factors to enhance silkworm defenses against invasive fungi. Insect Biochem. Mol. Biol. 2015, 57, 11–19. [Google Scholar] [CrossRef]
- Li, Y.S.; Liu, H.W.; Zhu, R.; Xia, Q.Y.; Zhao, P. Protease inhibitors in Bombyx mori silk might participate in protecting the pupating larva from microbial infection. Insect Sci. 2016, 23, 835–842. [Google Scholar] [CrossRef]
- Guo, X.; Dong, Z.; Zhang, Y.; Li, Y.; Liu, H.; Xia, Q.; Zhao, P. Proteins in the Cocoon of Silkworm Inhibit the Growth of Beauveria bassiana. PLoS ONE 2016, 11, e0151764. [Google Scholar] [CrossRef]
- Li, Y.; Liu, H.; Zhu, R.; Xia, Q.; Zhao, P. Loss of second and sixth conserved cysteine residues from trypsin inhibitor-like cysteine-rich domain-type protease inhibitors in Bombyx mori may induce activity against microbial proteases. Peptides 2016, 86, 13–23. [Google Scholar] [CrossRef]
- Li, Y.; Dong, Z.; Liu, H.; Zhu, R.; Bai, Y.; Xia, Q.; Zhao, P. The fungal-resistance factors BmSPI38 and BmSPI39 predominantly exist as tetramers, not monomers, in Bombyx mori. Insect Mol. Biol. 2018, 27, 686–697. [Google Scholar] [CrossRef]
- Luo, Z.; Yang, J.; Zhang, J.; Meng, G.; Lu, Q.; Yang, X.; Zhao, P.; Li, Y. Physicochemical Properties and Elimination of the Activity of Anti-Nutritional Serine Protease Inhibitors from Mulberry Leaves. Molecules 2022, 27, 1820. [Google Scholar]
- Jongsma, M.A.; Bakker, P.L.; Stiekema, W.J. Quantitative determination of serine proteinase inhibitor activity using a radial diffusion assay. Anal. Biochem. 1993, 212, 79–84. [Google Scholar]
- Yakoby, N.; Raskin, I. A simple method to determine trypsin and chymotrypsin inhibitory activity. J. Biochem. Biophys. Methods 2004, 59, 241–251. [Google Scholar] [CrossRef]
- Wriggers, W.; Chakravarty, S.; Jennings, P.A. Control of protein functional dynamics by peptide linkers. Biopolymers 2005, 80, 736–746. [Google Scholar]
- Zhao, H.L.; Yao, X.Q.; Xue, C.; Wang, Y.; Xiong, X.H.; Liu, Z.M. Increasing the homogeneity, stability and activity of human serum albumin and interferon-alpha2b fusion protein by linker engineering. Protein Expr. Purif. 2008, 61, 73–77. [Google Scholar] [CrossRef]
- Macedo, M.L.; Diz Filho, E.B.; Freire, M.G.; Oliva, M.L.; Sumikawa, J.T.; Toyama, M.H.; Marangoni, S. A trypsin inhibitor from Sapindus saponaria L. seeds: Purification, characterization, and activity towards pest insect digestive enzyme. Protein J. 2011, 30, 9–19. [Google Scholar] [CrossRef]
- Downing, M.R.; Bloom, J.W.; Mann, K.G. Comparison of the inhibition of thrombin by three plasma protease inhibitors. Biochemistry 1978, 17, 2649–2653. [Google Scholar] [CrossRef]
- Ellis, V.; Skully, M.; Macgregor, I.; Kakkar, V. Inhibition of human factor Xa by various plasma protease inhibitors. Biochim. Biophys. Acta 1982, 701, 24–31. [Google Scholar] [CrossRef]
- Zou, Z.; Jiang, H. Manduca sexta serpin-6 regulates immune serine proteinases PAP-3 and HP8. cDNA cloning, protein expression, inhibition kinetics, and function elucidation. J. Biol. Chem. 2005, 280, 14341–14348. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Kambris, Z.; Lemaitre, B.; Hashimoto, C. A serpin that regulates immune melanization in the respiratory system of Drosophila. Dev. Cell 2008, 15, 617–626. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Zhang, J.; Li, Y. Advances in the effect of multimerization of protease inhibitors on their physiological functions. Prog. Physiol. Sci. 2021, 52, 236–240. [Google Scholar]
- Gillmor, S.A.; Takeuchi, T.; Yang, S.Q.; Craik, C.S.; Fletterick, R.J. Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases. J. Mol. Biol. 2000, 299, 993–1003. [Google Scholar] [CrossRef]
- Nagy, Z.A.; Szakacs, D.; Boros, E.; Heja, D.; Vigh, E.; Sandor, N.; Jozsi, M.; Oroszlan, G.; Dobo, J.; Gal, P.; et al. Ecotin, a microbial inhibitor of serine proteases, blocks multiple complement dependent and independent microbicidal activities of human serum. PLoS Pathog. 2019, 15, e1008232. [Google Scholar] [CrossRef]
- Garcia, F.B.; Cabral, A.D.; Fuhlendorf, M.M.; da Cruz, G.F.; Dos Santos, J.V.; Ferreira, G.C.; de Rezende, B.R.C.; Santana, C.M.; Puzer, L.; Sasaki, S.D.; et al. Functional and structural characterization of an ecotin-like serine protease inhibitor from Trypanosoma cruzi. Int. J. Biol. Macromol. 2020, 151, 459–466. [Google Scholar] [CrossRef]
- Ekiel, I.; Abrahamson, M. Folding-related dimerization of human cystatin C. J. Biol. Chem. 1996, 271, 1314–1321. [Google Scholar] [CrossRef] [Green Version]
- Ekiel, I.; Abrahamson, M.; Fulton, D.B.; Lindahl, P.; Storer, A.C.; Levadoux, W.; Lafrance, M.; Labelle, S.; Pomerleau, Y.; Groleau, D.; et al. NMR structural studies of human cystatin C dimers and monomers. J. Mol. Biol. 1997, 271, 266–277. [Google Scholar] [CrossRef]
- Brand, G.D.; Salbo, R.; Jorgensen, T.J.; Bloch, C., Jr.; Boeri Erba, E.; Robinson, C.V.; Tanjoni, I.; Moura-da-Silva, A.M.; Roepstorff, P.; Domont, G.B.; et al. The interaction of the antitoxin DM43 with a snake venom metalloproteinase analyzed by mass spectrometry and surface plasmon resonance. J. Mass Spectrom. 2012, 47, 567–573. [Google Scholar] [CrossRef]
- Chapeaurouge, A.; Martins, S.M.; Holub, O.; Rocha, S.L.; Valente, R.H.; Neves-Ferreira, A.G.; Ferreira, S.T.; Domont, G.B.; Perales, J. Conformational plasticity of DM43, a metalloproteinase inhibitor from Didelphis marsupialis: Chemical and pressure-induced equilibrium (un)folding studies. Biochim Biophys Acta 2009, 1794, 1379–1386. [Google Scholar] [CrossRef]
- Wang, X.; Yang, G.; Li, S.; Gao, M.; Zhao, P.; Zhao, L. The Escherichia coli-derived thymosin beta4 concatemer promotes cell proliferation and healing wound in mice. Biomed. Res. Int. 2013, 2013, 241721. [Google Scholar]
- Chen, Y.; Zhao, L.; Shen, G.; Cui, L.; Ren, W.; Zhang, H.; Qian, H.; Tang, K. Expression and analysis of thymosin alpha1 concatemer in Escherichia coli. Biotechnol. Appl. Biochem. 2008, 49, 51–56. [Google Scholar]
- Fida, H.M.; Kumada, Y.; Terashima, M.; Katsuda, T.; Katoh, S. Tandem multimer expression of angiotensin I-converting enzyme inhibitory peptide in Escherichia coli. Biotechnol. J. 2009, 4, 1345–1356. [Google Scholar] [CrossRef]
- Park, C.J.; Lee, J.H.; Hong, S.S.; Lee, H.S.; Kim, S.C. High-level expression of the angiotensin-converting-enzyme-inhibiting peptide, YG-1, as tandem multimers in Escherichia coli. Appl. Microbiol. Biotechnol. 1998, 50, 71–76. [Google Scholar] [CrossRef]
- Liu, D.; Sun, H.; Zhang, L.; Li, S.; Qin, Z. High-level expression of milk-derived antihypertensive peptide in Escherichia coli and its bioactivity. J. Agric. Food Chem. 2007, 55, 5109–5112. [Google Scholar] [CrossRef]
- Rao, X.; Hu, J.; Li, S.; Jin, X.; Zhang, C.; Cong, Y.; Hu, X.; Tan, Y.; Huang, J.; Chen, Z.; et al. Design and expression of peptide antibiotic hPAB-beta as tandem multimers in Escherichia coli. Peptides 2005, 26, 721–729. [Google Scholar] [CrossRef]
- Wang, F.J.; Song, H.L.; Wang, X.M.; Zhang, W.J.; Wang, B.L.; Zhao, J.; Hu, Z.B. Tandem multimer expression and preparation of hypoglycemic peptide MC6 from Momordica charantia in Escherichia coli. Appl. Biochem. Biotechnol. 2012, 166, 612–619. [Google Scholar] [CrossRef]
- Guler, H.I. Recombinant production of Opiorphin pentapeptide as tandem multimers through rational design of primers. Appl. Biochem. Microbiol. 2020, 56, 141–148. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, K.; Dong, Z.; Chen, Z.; Zhu, H.; Zhang, Y.; Xia, Q.; Zhao, P. Kunitz-type protease inhibitor BmSPI51 plays an antifungal role in the silkworm cocoon. Insect Biochem. Mol. Biol. 2020, 116, 103258. [Google Scholar]
- Chen, Z.Y.; Brown, R.L.; Lax, A.R.; Cleveland, T.E.; Russin, J.S. Inhibition of plant-pathogenic fungi by a corn trypsin inhibitor overexpressed in Escherichia coli. Appl. Env. Microbiol. 1999, 65, 1320–1324. [Google Scholar] [CrossRef] [Green Version]






| Primers | Sequence (5′ → 3′) |
|---|---|
| BmSPI39-Nde I-BamH I-F | CGCCATATGGGCGGATCCTTTGAAAAAGATTGTCCTGAGAATTCT |
| BmSPI39-Not I-R | ATTTGCGGCCGCTTATGACTGTTGTTTATGGAAACAGTTG |
| BmSPI39-Bgl II-R | GAAGATCTTGACTGTTGTTTATGGAAACAGTTGAC |
| BmSPI39-L-Bgl II-R | GAAGATCTTGAGCCACCACCGCCTGAGCCACCACCGCCTGAGCC ACCACCGCCTGACTGTTGTTTATGGAAACAGTTGAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Wang, Y.; Zhu, R.; Yang, X.; Wei, M.; Zhang, Z.; Chen, C.; Zhao, P. Tandem Multimerization Can Enhance the Structural Homogeneity and Antifungal Activity of the Silkworm Protease Inhibitor BmSPI39. Cells 2023, 12, 693. https://doi.org/10.3390/cells12050693
Li Y, Wang Y, Zhu R, Yang X, Wei M, Zhang Z, Chen C, Zhao P. Tandem Multimerization Can Enhance the Structural Homogeneity and Antifungal Activity of the Silkworm Protease Inhibitor BmSPI39. Cells. 2023; 12(5):693. https://doi.org/10.3390/cells12050693
Chicago/Turabian StyleLi, Youshan, Yuan Wang, Rui Zhu, Xi Yang, Meng Wei, Zhaofeng Zhang, Changqing Chen, and Ping Zhao. 2023. "Tandem Multimerization Can Enhance the Structural Homogeneity and Antifungal Activity of the Silkworm Protease Inhibitor BmSPI39" Cells 12, no. 5: 693. https://doi.org/10.3390/cells12050693
APA StyleLi, Y., Wang, Y., Zhu, R., Yang, X., Wei, M., Zhang, Z., Chen, C., & Zhao, P. (2023). Tandem Multimerization Can Enhance the Structural Homogeneity and Antifungal Activity of the Silkworm Protease Inhibitor BmSPI39. Cells, 12(5), 693. https://doi.org/10.3390/cells12050693