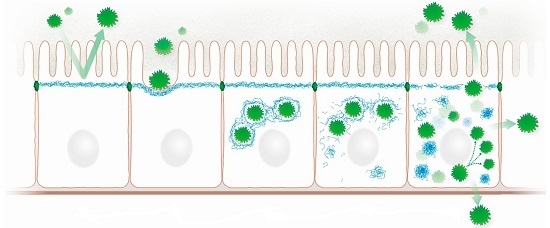
Epithelial Intermediate Filaments: Guardians against Microbial Infection?
Abstract
:1. Intermediate Filaments: Organization and Function
2. Barrier Function of Intermediate Filaments in Stratified Epithelia
3. Keratin-Microbe Interactions in Stratified Epithelia
3.1. Epithelial Colonization
3.2. Keratin Network Disruption
3.3. Induction of Inflammation
3.4. Bacteriotoxicity
4. Intermediate Filaments Mediating Barrier Function in Simple Epithelia
5. Pathogens Interfering with Barrier Function in Simple Epithelia
5.1. Pathogen Docking
5.2. Induction of Cytotoxic Effects
5.3. Keratin Network Disruption
5.4. Pathogen Proliferation and Survival
6. The C. elegans Intestine as a Model System for Investigating Intermediate Filament-Microbe Interactions
7. Conclusions and Outlook
Acknowledgments
Conflicts of Interest
References
- Kim, S.; Coulombe, P.A. Intermediate filament scaffolds fulfill mechanical, organizational, and signaling functions in the cytoplasm. Genes Dev. 2007, 21, 1581–1597. [Google Scholar] [CrossRef] [PubMed]
- Magin, T.M.; Vijayaraj, P.; Leube, R.E. Structural and regulatory functions of keratins. Exp. Cell Res. 2007, 313, 2021–2032. [Google Scholar] [CrossRef] [PubMed]
- Toivola, D.M.; Strnad, P.; Habtezion, A.; Omary, M.B. Intermediate filaments take the heat as stress proteins. Trends Cell Biol. 2010, 20, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Margiotta, A.; Bucci, C. Role of intermediate filaments in vesicular traffic. Cells 2016, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Bar, H.; Kreplak, L.; Strelkov, S.V.; Aebi, U. Intermediate filaments: From cell architecture to nanomechanics. Nat. Rev. Mol. Cell Biol. 2007, 8, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Godsel, L.M.; Hobbs, R.P.; Green, K.J. Intermediate filament assembly: Dynamics to disease. Trends Cell Biol. 2008, 18, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Goldman, R.D.; Grin, B.; Mendez, M.G.; Kuczmarski, E.R. Intermediate filaments: Versatile building blocks of cell structure. Curr. Opin. Cell Biol. 2008, 20, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Strelkov, S.V.; Burkhard, P.; Aebi, U. Intermediate filaments: Primary determinants of cell architecture and plasticity. J. Clin. Investig. 2009, 119, 1772–1783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Windoffer, R.; Beil, M.; Magin, T.M.; Leube, R.E. Cytoskeleton in motion: The dynamics of keratin intermediate filaments in epithelia. J. Cell Biol. 2011, 194, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstern, T.; Mucke, N.; Aebi, U.; Mauermann, M.; Herrmann, H. Complex formation and kinetics of filament assembly exhibited by the simple epithelial keratins K8 and K18. J. Struct. Biol. 2012, 177, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, H.; Suda, M. Seven kinds of intermediate filament networks in the cytoplasm of polarized cells: Structure and function. Acta Histochem. Cytochem. 2010, 43, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Leube, R.E.; Schwarz, N. Intermediate filaments. In Encyclopedia of Cell Biology; Bradshaw, R.A., Stahl, P.D., Eds.; Academic Press: Waltham, MA, USA, 2016; Volume 2, pp. 569–578. [Google Scholar]
- Coulombe, P.A.; Wong, P. Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat. Cell Biol. 2004, 6, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Stamenovic, D.; Wang, N. Invited review: Engineering approaches to cytoskeletal mechanics. J. Appl. Physiol. 2000, 89, 2085–2090. [Google Scholar] [PubMed]
- Ramms, L.; Fabris, G.; Windoffer, R.; Schwarz, N.; Springer, R.; Zhou, C.; Lazar, J.; Stiefel, S.; Hersch, N.; Schnakenberg, U.; et al. Keratins as the main component for the mechanical integrity of keratinocytes. Proc. Natl. Acad. Sci. USA 2013, 110, 18513–18518. [Google Scholar] [CrossRef] [PubMed]
- Seltmann, K.; Fritsch, A.W.; Kas, J.A.; Magin, T.M. Keratins significantly contribute to cell stiffness and impact invasive behavior. Proc. Natl. Acad. Sci. USA 2013, 110, 18507–18512. [Google Scholar] [CrossRef] [PubMed]
- Mendez, M.G.; Restle, D.; Janmey, P.A. Vimentin enhances cell elastic behavior and protects against compressive stress. Biophys. J. 2014, 107, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Homberg, M.; Ramms, L.; Schwarz, N.; Dreissen, G.; Leube, R.E.; Merkel, R.; Hoffmann, B.; Magin, T.M. Distinct impact of two keratin mutations causing epidermolysis bullosa simplex on keratinocyte adhesion and stiffness. J. Investig. Dermatol. 2015, 135, 2437–2445. [Google Scholar] [CrossRef] [PubMed]
- Kroger, C.; Loschke, F.; Schwarz, N.; Windoffer, R.; Leube, R.E.; Magin, T.M. Keratins control intercellular adhesion involving PKC-alpha-mediated desmoplakin phosphorylation. J. Cell Biol. 2013, 201, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Omary, M.B.; Coulombe, P.A.; McLean, W.H. Intermediate filament proteins and their associated diseases. N. Engl. J. Med. 2004, 351, 2087–2100. [Google Scholar] [CrossRef] [PubMed]
- Omary, M.B.; Ku, N.O.; Strnad, P.; Hanada, S. Toward unraveling the complexity of simple epithelial keratins in human disease. J. Clin. Investig. 2009, 119, 1794–1805. [Google Scholar] [CrossRef] [PubMed]
- Chamcheu, J.C.; Siddiqui, I.A.; Syed, D.N.; Adhami, V.M.; Liovic, M.; Mukhtar, H. Keratin gene mutations in disorders of human skin and its appendages. Arch. Biochem. Biophys. 2011, 508, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Homberg, M.; Magin, T.M. Beyond expectations: Novel insights into epidermal keratin function and regulation. Int. Rev. Cell Mol. Biol. 2014, 311, 265–306. [Google Scholar] [PubMed]
- Chamcheu, J.C.; Navsaria, H.; Pihl-Lundin, I.; Liovic, M.; Vahlquist, A.; Torma, H. Chemical chaperones protect epidermolysis bullosa simplex keratinocytes from heat stress-induced keratin aggregation: Involvement of heat shock proteins and map kinases. J. Investig. Dermatol. 2011, 131, 1684–1691. [Google Scholar] [CrossRef] [PubMed]
- Werner, N.S.; Windoffer, R.; Strnad, P.; Grund, C.; Leube, R.E.; Magin, T.M. Epidermolysis bullosa simplex-type mutations alter the dynamics of the keratin cytoskeleton and reveal a contribution of actin to the transport of keratin subunits. Mol. Biol. Cell 2004, 15, 990–1002. [Google Scholar] [CrossRef] [PubMed]
- Wöll, S.; Windoffer, R.; Leube, R.E. p38 MAPK-dependent shaping of the keratin cytoskeleton in cultured cells. J. Cell Biol. 2007, 177, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Owens, D.W.; Lane, E.B. Keratin mutations and intestinal pathology. J. Pathol. 2004, 204, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Ku, N.O.; Lim, J.K.; Krams, S.M.; Esquivel, C.O.; Keeffe, E.B.; Wright, T.L.; Parry, D.A.; Omary, M.B. Keratins as susceptibility genes for end-stage liver disease. Gastroenterology 2005, 129, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Hertel, L. Herpesviruses and intermediate filaments: Close encounters with the third type. Viruses 2011, 3, 1015–1040. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J. The E4 protein; structure, function and patterns of expression. Virology 2013, 445, 80–98. [Google Scholar] [CrossRef] [PubMed]
- Haglund, C.M.; Welch, M.D. Pathogens and polymers: Microbe-host interactions illuminate the cytoskeleton. J. Cell Biol. 2011, 195, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Mak, T.N.; Bruggemann, H. Vimentin in bacterial infections. Cells 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Owens, D.W.; Lane, E.B. The quest for the function of simple epithelial keratins. Bioessays 2003, 25, 748–758. [Google Scholar] [CrossRef] [PubMed]
- Irvine, A.D.; McLean, W.H. Human keratin diseases: The increasing spectrum of disease and subtlety of the phenotype-genotype correlation. Br. J. Dermatol. 1999, 140, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Leachman, S.A.; Kaspar, R.L.; Fleckman, P.; Florell, S.R.; Smith, F.J.; McLean, W.H.; Lunny, D.P.; Milstone, L.M.; van Steensel, M.A.; Munro, C.S.; et al. Clinical and pathological features of pachyonychia congenita. J. Investig. Dermatol. Symp. Proc. 2005, 10, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Schmuth, M.; Yosipovitch, G.; Williams, M.L.; Weber, F.; Hintner, H.; Ortiz-Urda, S.; Rappersberger, K.; Crumrine, D.; Feingold, K.R.; Elias, P.M. Pathogenesis of the permeability barrier abnormality in epidermolytic hyperkeratosis. J. Investig. Dermatol. 2001, 117, 837–847. [Google Scholar] [CrossRef] [PubMed]
- Lane, E.B.; McLean, W.H. Keratins and skin disorders. J. Pathol. 2004, 204, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Segre, J.A. Epidermal barrier formation and recovery in skin disorders. J. Clin. Investig. 2006, 116, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Arin, M.J.; Oji, V.; Emmert, S.; Hausser, I.; Traupe, H.; Krieg, T.; Grimberg, G. Expanding the keratin mutation database: Novel and recurrent mutations and genotype-phenotype correlations in 28 patients with epidermolytic ichthyosis. Br. J. Dermatol. 2011, 164, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Vijayaraj, P.; Sohl, G.; Magin, T.M. Keratin transgenic and knockout mice: Functional analysis and validation of disease-causing mutations. Methods Mol. Biol. 2007, 360, 203–251. [Google Scholar] [PubMed]
- Jensen, J.M.; Schutze, S.; Neumann, C.; Proksch, E. Impaired cutaneous permeability barrier function, skin hydration, and sphingomyelinase activity in keratin 10 deficient mice. J. Investig. Dermatol. 2000, 115, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Bouameur, J.E.; Bar, J.; Rice, R.H.; Hornig-Do, H.T.; Roop, D.R.; Schwarz, N.; Brodesser, S.; Thiering, S.; Leube, R.E.; et al. A keratin scaffold regulates epidermal barrier formation, mitochondrial lipid composition, and activity. J. Cell Biol. 2015, 211, 1057–1075. [Google Scholar] [CrossRef] [PubMed]
- Liovic, M.; D’Alessandro, M.; Tomic-Canic, M.; Bolshakov, V.N.; Coats, S.E.; Lane, E.B. Severe keratin 5 and 14 mutations induce down-regulation of junction proteins in keratinocytes. Exp. Cell Res. 2009, 315, 2995–3003. [Google Scholar] [CrossRef] [PubMed]
- DiTommaso, T.; Cottle, D.L.; Pearson, H.B.; Schluter, H.; Kaur, P.; Humbert, P.O.; Smyth, I.M. Keratin 76 is required for tight junction function and maintenance of the skin barrier. PLoS Genet. 2014, 10, e1004706. [Google Scholar] [CrossRef] [PubMed]
- Wallace, L.; Roberts-Thompson, L.; Reichelt, J. Deletion of K1/K10 does not impair epidermal stratification but affects desmosomal structure and nuclear integrity. J. Cell Sci. 2012, 125, 1750–1758. [Google Scholar] [CrossRef] [PubMed]
- Lessard, J.C.; Pina-Paz, S.; Rotty, J.D.; Hickerson, R.P.; Kaspar, R.L.; Balmain, A.; Coulombe, P.A. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc. Natl. Acad. Sci. USA 2013, 110, 19537–19542. [Google Scholar] [CrossRef] [PubMed]
- Roth, W.; Kumar, V.; Beer, H.D.; Richter, M.; Wohlenberg, C.; Reuter, U.; Thiering, S.; Staratschek-Jox, A.; Hofmann, A.; Kreusch, F.; et al. Keratin 1 maintains skin integrity and participates in an inflammatory network in skin through interleukin-18. J. Cell Sci. 2012, 125, 5269–5279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, L.; Wang, G. Keratin 17: A critical player in the pathogenesis of psoriasis. Med. Res. Rev. 2014, 34, 438–454. [Google Scholar] [CrossRef] [PubMed]
- Depianto, D.; Kerns, M.L.; Dlugosz, A.A.; Coulombe, P.A. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat. Genet. 2010, 42, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, R.P.; DePianto, D.J.; Jacob, J.T.; Han, M.C.; Chung, B.M.; Batazzi, A.S.; Poll, B.G.; Guo, Y.; Han, J.; Ong, S.; et al. Keratin-dependent regulation of aire and gene expression in skin tumor keratinocytes. Nat. Genet. 2015, 47, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.M.; Arutyunov, A.; Ilagan, E.; Yao, N.; Wills-Karp, M.; Coulombe, P.A. Regulation of C-X-C chemokine gene expression by keratin 17 and hnRNP K in skin tumor keratinocytes. J. Cell Biol. 2015, 208, 613–627. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, L.M.; Walsh, E.J.; Massey, R.C.; Peacock, S.J.; Foster, T.J. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: Implications for nasal colonization. Cell Microbiol. 2002, 4, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Samen, U.; Eikmanns, B.J.; Reinscheid, D.J.; Borges, F. The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect. Immun. 2007, 75, 5405–5414. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, P.B.; Laskey, P.; Sullivan, K.; Davy, C.; Wang, Q.; Jackson, D.J.; Griffin, H.M.; Doorbar, J. E1–E4-mediated keratin phosphorylation and ubiquitylation: A mechanism for keratin depletion in HPV16-infected epithelium. J. Cell Sci. 2010, 123, 2810–2822. [Google Scholar] [CrossRef] [PubMed]
- Goshima, F.; Watanabe, D.; Suzuki, H.; Takakuwa, H.; Yamada, H.; Nishiyama, Y. The US2 gene product of herpes simplex virus type 2 interacts with cytokeratin 18. Arch. Virol. 2001, 146, 2201–2209. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Goshima, F.; Nishizawa, Y.; Daikoku, T.; Takakuwa, H.; Ohtsuka, K.; Yoshikawa, T.; Nishiyama, Y. Phosphorylation of cytokeratin 17 by herpes simplex virus type 2 US3 protein kinase. Microbiol. Immunol. 2002, 46, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Tancharoen, S.; Matsuyama, T.; Kawahara, K.; Tanaka, K.; Lee, L.J.; Machigashira, M.; Noguchi, K.; Ito, T.; Imamura, T.; Potempa, J.; et al. Cleavage of host cytokeratin-6 by lysine-specific gingipain induces gingival inflammation in periodontitis patients. PLoS ONE 2015, 10, e0117775. [Google Scholar] [CrossRef] [PubMed]
- Tam, C.; Mun, J.J.; Evans, D.J.; Fleiszig, S.M. Cytokeratins mediate epithelial innate defense through their antimicrobial properties. J. Clin. Investig. 2012, 122, 3665–3677. [Google Scholar] [CrossRef] [PubMed]
- Batchelor, M.; Guignot, J.; Patel, A.; Cummings, N.; Cleary, J.; Knutton, S.; Holden, D.W.; Connerton, I.; Frankel, G. Involvement of the intermediate filament protein cytokeratin-18 in actin pedestal formation during EPEC infection. EMBO Rep. 2004, 5, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Carlson, S.A.; Omary, M.B.; Jones, B.D. Identification of cytokeratins as accessory mediators of Salmonella entry into eukaryotic cells. Life Sci. 2002, 70, 1415–1426. [Google Scholar] [CrossRef]
- Scherer, C.A.; Cooper, E.; Miller, S.I. The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol. Microbiol. 2000, 37, 1133–1145. [Google Scholar] [CrossRef] [PubMed]
- Russo, B.C.; Stamm, L.M.; Raaben, M.; Kim, C.M.; Kahoud, E.; Robinson, L.R.; Bose, S.; Queiroz, A.L.; Herrera, B.B.; Baxt, L.A.; et al. Intermediate filaments enable pathogen docking to trigger type 3 effector translocation. Nat. Microbiol. 2016, 1, 16025. [Google Scholar] [CrossRef]
- Nava-Acosta, R.; Navarro-Garcia, F. Cytokeratin 8 is an epithelial cell receptor for pet, a cytotoxic serine protease autotransporter of enterobacteriaceae. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Lowthert, L.A.; Omary, M.B. Heat stress or rotavirus infection of human epithelial cells generates a distinct hyperphosphorylated form of keratin 8. Exp. Cell Res. 1995, 219, 348–357. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.H.; Ornelles, D.A.; Shenk, T. The adenovirus L3 23-kilodalton proteinase cleaves the amino-terminal head domain from cytokeratin 18 and disrupts the cytokeratin network of hela cells. J. Virol. 1993, 67, 3507–3514. [Google Scholar] [PubMed]
- Zhang, Y.; Schneider, R.J. Adenovirus inhibition of cell translation facilitates release of virus particles and enhances degradation of the cytokeratin network. J. Virol. 1994, 68, 2544–2555. [Google Scholar] [PubMed]
- Seipelt, J.; Liebig, H.D.; Sommergruber, W.; Gerner, C.; Kuechler, E. 2A proteinase of human rhinovirus cleaves cytokeratin 8 in infected HeLa cells. J. Biol. Chem. 2000, 275, 20084–20089. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Su, H.; Huang, Y.; Zhong, Y.; Zhong, G. Cleavage of host keratin 8 by a chlamydia-secreted protease. Infect. Immun. 2004, 72, 3863–3868. [Google Scholar] [CrossRef] [PubMed]
- Savijoki, K.; Alvesalo, J.; Vuorela, P.; Leinonen, M.; Kalkkinen, N. Proteomic analysis of Chlamydia pneumoniae-infected HL cells reveals extensive degradation of cytoskeletal proteins. FEMS Immunol. Med. Microbiol. 2008, 54, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.A.; de Vasconcelos Vde, C.; Colli, W.; Alves, M.J.; Giordano, R.J. Trypanosoma cruzi binds to cytokeratin through conserved peptide motifs found in the laminin-g-like domain of the gp85/trans-sialidase proteins. PLoS Negl. Trop. Dis. 2015, 9, e0004099. [Google Scholar] [CrossRef] [PubMed]
- Weidner, E.; Halonen, S.K. Microsporidian spore envelope keratins phosphorylate and disassemble during spore activation. J. Eukaryot. Microbiol. 1993, 40, 783–788. [Google Scholar] [CrossRef]
- Ganesh, V.K.; Barbu, E.M.; Deivanayagam, C.C.; Le, B.; Anderson, A.S.; Matsuka, Y.V.; Lin, S.L.; Foster, T.J.; Narayana, S.V.; Hook, M. Structural and biochemical characterization of Staphylococcus aureus clumping factor B/ligand interactions. J. Biol. Chem. 2011, 286, 25963–25972. [Google Scholar] [CrossRef] [PubMed]
- Walsh, E.J.; O'Brien, L.M.; Liang, X.; Hook, M.; Foster, T.J. Clumping factor B, a fibrinogen-binding mscramm (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10. J. Biol. Chem. 2004, 279, 50691–50699. [Google Scholar] [CrossRef] [PubMed]
- Bragulla, H.H.; Homberger, D.G. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. J. Anat. 2009, 214, 516–559. [Google Scholar] [CrossRef] [PubMed]
- Moll, R.; Divo, M.; Langbein, L. The human keratins: Biology and pathology. Histochem. Cell Biol. 2008, 129, 705–733. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, M.E.; Geoghegan, J.A.; Monk, I.R.; O’Keeffe, K.M.; Walsh, E.J.; Foster, T.J.; McLoughlin, R.M. Nasal colonisation by Staphylococcus aureus depends upon clumping factor B binding to the squamous epithelial cell envelope protein loricrin. PLoS Pathog. 2012, 8, e1003092. [Google Scholar] [CrossRef] [PubMed]
- Haim, M.; Trost, A.; Maier, C.J.; Achatz, G.; Feichtner, S.; Hintner, H.; Bauer, J.W.; Onder, K. Cytokeratin 8 interacts with clumping factor B: A new possible virulence factor target. Microbiology 2010, 156, 3710–3721. [Google Scholar] [CrossRef] [PubMed]
- Tamura, G.S.; Nittayajarn, A. Group B streptococci and other gram-positive cocci bind to cytokeratin 8. Infect. Immun. 2000, 68, 2129–2134. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Ely, S.; Sterling, J.; McLean, C.; Crawford, L. Specific interaction between HPV-16 E1–E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 1991, 352, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Khan, J.; Davy, C.E.; McIntosh, P.B.; Jackson, D.J.; Hinz, S.; Wang, Q.; Doorbar, J. Role of calpain in the formation of human papillomavirus type 16 E1^E4 amyloid fibers and reorganization of the keratin network. J. Virol. 2011, 85, 9984–9997. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Griffin, H.; Southern, S.; Jackson, D.; Martin, A.; McIntosh, P.; Davy, C.; Masterson, P.J.; Walker, P.A.; Laskey, P.; et al. Functional analysis of the human papillomavirus type 16 E1=E4 protein provides a mechanism for in vivo and in vitro keratin filament reorganization. J. Virol. 2004, 78, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Kennedy, A.; Das, P.; McIntosh, P.B.; Howell, S.A.; Isaacson, E.R.; Hinz, S.A.; Davy, C.; Doorbar, J. Phosphorylation of the human papillomavirus type 16 E1–E4 protein at T57 by ERK triggers a structural change that enhances keratin binding and protein stability. J. Virol. 2009, 83, 3668–3683. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.H.; Roy, B.B.; Finnen, R.L.; Le Sage, V.; Johnston, S.M.; Zhang, H.; Banfield, B.W. The US2 gene product of herpes simplex virus 2 is a membrane-associated ubiquitin-interacting protein. J. Virol. 2013, 87, 9590–9603. [Google Scholar] [CrossRef] [PubMed]
- Lyman, M.G.; Randall, J.A.; Calton, C.M.; Banfield, B.W. Localization of ERK/MAP kinase is regulated by the alphaherpesvirus tegument protein US2. J. Virol. 2006, 80, 7159–7168. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Goshima, F.; Daikoku, T.; Takakuwa, H.; Nishiyama, Y. Expression of herpes simplex virus type 2 US3 affects the Cdc42/Rac pathway and attenuates c-Jun N-terminal kinase activation. Genes Cells 2000, 5, 1017–1027. [Google Scholar] [CrossRef] [PubMed]
- Imamura, T. The role of gingipains in the pathogenesis of periodontal disease. J. Periodontol. 2003, 74, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Grenier, D.; Imbeault, S.; Plamondon, P.; Grenier, G.; Nakayama, K.; Mayrand, D. Role of gingipains in growth of porphyromonas gingivalis in the presence of human serum albumin. Infect. Immun. 2001, 69, 5166–5172. [Google Scholar] [CrossRef] [PubMed]
- Zasloff, M. Defending the cornea with antibacterial fragments of keratin. J. Clin. Investig. 2012, 122, 3471–3473. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, N.; Windoffer, R.; Magin, T.M.; Leube, R.E. Dissection of keratin network formation, turnover and reorganization in living murine embryos. Sci. Rep. 2015, 5, 9007. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Toivola, D.M.; Feng, N.; Greenberg, H.B.; Franke, W.W.; Omary, M.B. Keratin 20 helps maintain intermediate filament organization in intestinal epithelia. Mol. Biol. Cell 2003, 14, 2959–2971. [Google Scholar] [CrossRef] [PubMed]
- Toivola, D.M.; Baribault, H.; Magin, T.; Michie, S.A.; Omary, M.B. Simple epithelial keratins are dispensable for cytoprotection in two pancreatitis models. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 279, G1343–G1354. [Google Scholar] [PubMed]
- Franke, W.W.; Appelhans, B.; Schmid, E.; Freudenstein, C.; Osborn, M.; Weber, K. The organization of cytokeratin filaments in the intestinal epithelium. Eur. J. Cell Biol. 1979, 19, 255–268. [Google Scholar] [PubMed]
- Moll, R.; Schiller, D.L.; Franke, W.W. Identification of protein it of the intestinal cytoskeleton as a novel type I cytokeratin with unusual properties and expression patterns. J. Cell Biol. 1990, 111, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Moll, R.; Zimbelmann, R.; Goldschmidt, M.D.; Keith, M.; Laufer, J.; Kasper, M.; Koch, P.J.; Franke, W.W. The human gene encoding cytokeratin 20 and its expression during fetal development and in gastrointestinal carcinomas. Differentiation 1993, 53, 75–93. [Google Scholar] [CrossRef] [PubMed]
- Djudjaj, S.; Papasotiriou, M.; Bulow, R.D.; Wagnerova, A.; Lindenmeyer, M.T.; Cohen, C.D.; Strnad, P.; Goumenos, D.S.; Floege, J.; Boor, P. Keratins are novel markers of renal epithelial cell injury. Kidney Int. 2016, 89, 792–808. [Google Scholar] [CrossRef] [PubMed]
- Guldiken, N.; Ensari, G.K.; Lahiri, P.; Couchy, G.; Preisinger, C.; Liedtke, C.; Zimmermann, H.W.; Ziol, M.; Boor, P.; Zucman-Rossi, J.; et al. Keratin 23 is a general stress-inducible marker of mouse and human ductular reaction in liver disease. J. Hepatol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Guldiken, N.; Usachov, V.; Levada, K.; Trautwein, C.; Ziol, M.; Nahon, P.; Strnad, P. Keratins 8 and 18 are type II acute-phase responsive genes overexpressed in human liver disease. Liver Int. 2015, 35, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Zatloukal, K.; Stumptner, C.; Fuchsbichler, A.; Fickert, P.; Lackner, C.; Trauner, M.; Denk, H. The keratin cytoskeleton in liver diseases. J. Pathol. 2004, 204, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Ku, N.O.; Gish, R.; Wright, T.L.; Omary, M.B. Keratin 8 mutations in patients with cryptogenic liver disease. N. Engl. J. Med. 2001, 344, 1580–1587. [Google Scholar] [CrossRef] [PubMed]
- Ku, N.O.; Darling, J.M.; Krams, S.M.; Esquivel, C.O.; Keeffe, E.B.; Sibley, R.K.; Lee, Y.M.; Wright, T.L.; Omary, M.B. Keratin 8 and 18 mutations are risk factors for developing liver disease of multiple etiologies. Proc. Natl. Acad. Sci. USA 2003, 100, 6063–6068. [Google Scholar] [CrossRef] [PubMed]
- Cavestro, G.M.; Frulloni, L.; Nouvenne, A.; Neri, T.M.; Calore, B.; Ferri, B.; Bovo, P.; Okolicsanyi, L.; di Mario, F.; Cavallini, G. Association of keratin 8 gene mutation with chronic pancreatitis. Dig. Liver Dis. 2003, 35, 416–420. [Google Scholar] [CrossRef]
- Owens, D.W.; Wilson, N.J.; Hill, A.J.; Rugg, E.L.; Porter, R.M.; Hutcheson, A.M.; Quinlan, R.A.; van Heel, D.; Parkes, M.; Jewell, D.P.; et al. Human keratin 8 mutations that disturb filament assembly observed in inflammatory bowel disease patients. J. Cell Sci. 2004, 117, 1989–1999. [Google Scholar] [CrossRef] [PubMed]
- Toivola, D.M.; Boor, P.; Alam, C.; Strnad, P. Keratins in health and disease. Curr. Opin. Cell Biol. 2015, 32, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Tao, G.Z.; Strnad, P.; Zhou, Q.; Kamal, A.; Zhang, L.; Madani, N.D.; Kugathasan, S.; Brant, S.R.; Cho, J.H.; Omary, M.B.; et al. Analysis of keratin polypeptides 8 and 19 variants in inflammatory bowel disease. Clin. Gastroenterol. Hepatol. 2007, 5, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Treiber, M.; Schulz, H.U.; Landt, O.; Drenth, J.P.; Castellani, C.; Real, F.X.; Akar, N.; Ammann, R.W.; Bargetzi, M.; Bhatia, E.; et al. Keratin 8 sequence variants in patients with pancreatitis and pancreatic cancer. J. Mol. Med. (Berl.) 2006, 84, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Baribault, H.; Penner, J.; Iozzo, R.V.; Wilson-Heiner, M. Colorectal hyperplasia and inflammation in keratin 8-deficient FVB/N mice. Genes Dev. 1994, 8, 2964–2973. [Google Scholar] [CrossRef] [PubMed]
- Habtezion, A.; Toivola, D.M.; Butcher, E.C.; Omary, M.B. Keratin-8-deficient mice develop chronic spontaneous Th2 colitis amenable to antibiotic treatment. J. Cell Sci. 2005, 118, 1971–1980. [Google Scholar] [CrossRef] [PubMed]
- Misiorek, J.O.; Lahdeniemi, I.A.; Nystrom, J.H.; Paramonov, V.M.; Gullmets, J.A.; Saarento, H.; Rivero-Muller, A.; Husoy, T.; Taimen, P.; Toivola, D.M. Keratin 8-deletion induced colitis predisposes to murine colorectal cancer enforced by the inflammasome and IL-22 pathway. Carcinogenesis 2016. [Google Scholar] [CrossRef] [PubMed]
- Habtezion, A.; Toivola, D.M.; Asghar, M.N.; Kronmal, G.S.; Brooks, J.D.; Butcher, E.C.; Omary, M.B. Absence of keratin 8 confers a paradoxical microflora-dependent resistance to apoptosis in the colon. Proc. Natl. Acad. Sci. USA 2011, 108, 1445–1450. [Google Scholar] [CrossRef] [PubMed]
- Asghar, M.N.; Silvander, J.S.; Helenius, T.O.; Lahdeniemi, I.A.; Alam, C.; Fortelius, L.E.; Holmsten, R.O.; Toivola, D.M. The amount of keratins matters for stress protection of the colonic epithelium. PLoS ONE 2015, 10, e0127436. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, S.; Loranger, A.; Daigle, N.; Marceau, N. Simple epithelium keratins 8 and 18 provide resistance to Fas-mediated apoptosis. The protection occurs through a receptor-targeting modulation. J. Cell Biol. 2001, 154, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Caulin, C.; Ware, C.F.; Magin, T.M.; Oshima, R.G. Keratin-dependent, epithelial resistance to tumor necrosis factor-induced apoptosis. J. Cell Biol. 2000, 149, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Inada, H.; Izawa, I.; Nishizawa, M.; Fujita, E.; Kiyono, T.; Takahashi, T.; Momoi, T.; Inagaki, M. Keratin attenuates tumor necrosis factor-induced cytotoxicity through association with tradd. J. Cell Biol. 2001, 155, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, N.J.; Allen, R.E.; Stevens, T.R.; van Someren, R.N.; Blake, D.R.; Rampton, D.S. Chemiluminescence assay of mucosal reactive oxygen metabolites in inflammatory bowel disease. Gastroenterology 1992, 103, 186–196. [Google Scholar] [CrossRef]
- McKenzie, S.J.; Baker, M.S.; Buffinton, G.D.; Doe, W.F. Evidence of oxidant-induced injury to epithelial cells during inflammatory bowel disease. J. Clin. Investig. 1996, 98, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Keshavarzian, A.; Banan, A.; Farhadi, A.; Komanduri, S.; Mutlu, E.; Zhang, Y.; Fields, J.Z. Increases in free radicals and cytoskeletal protein oxidation and nitration in the colon of patients with inflammatory bowel disease. Gut 2003, 52, 720–728. [Google Scholar] [CrossRef] [PubMed]
- Corfe, B.M.; Majumdar, D.; Assadsangabi, A.; Marsh, A.M.; Cross, S.S.; Connolly, J.B.; Evans, C.A.; Lobo, A.J. Inflammation decreases keratin level in ulcerative colitis; inadequate restoration associates with increased risk of colitis-associated cancer. BMJ Open Gastroenterol. 2015, 2, e000024. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Srinivasan, S.; Theiss, A.L.; Merlin, D.; Sitaraman, S.V. Interleukin-6 induces keratin expression in intestinal epithelial cells: Potential role of keratin-8 in interleukin-6-induced barrier function alterations. J. Biol. Chem. 2007, 282, 8219–8227. [Google Scholar] [CrossRef] [PubMed]
- Fujino, S.; Andoh, A.; Bamba, S.; Ogawa, A.; Hata, K.; Araki, Y.; Bamba, T.; Fujiyama, Y. Increased expression of interleukin 17 in inflammatory bowel disease. Gut 2003, 52, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Zahs, A.; Bird, M.D.; Ramirez, L.; Choudhry, M.A.; Kovacs, E.J. Anti-IL-6 antibody treatment but not IL-6 knockout improves intestinal barrier function and reduces inflammation after binge ethanol exposure and burn injury. Shock 2013, 39, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Toivola, D.M.; Krishnan, S.; Binder, H.J.; Singh, S.K.; Omary, M.B. Keratins modulate colonocyte electrolyte transport via protein mistargeting. J. Cell Biol. 2004, 164, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Salas, P.J.; Rodriguez, M.L.; Viciana, A.L.; Vega-Salas, D.E.; Hauri, H.P. The apical submembrane cytoskeleton participates in the organization of the apical pole in epithelial cells. J. Cell Biol. 1997, 137, 359–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asghar, M.N.; Priyamvada, S.; Nystrom, J.H.; Anbazhagan, A.N.; Dudeja, P.K.; Toivola, D.M. Keratin 8 knockdown leads to loss of the chloride transporter DRA in the colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Helenius, T.O.; Misiorek, J.O.; Nystrom, J.H.; Fortelius, L.E.; Habtezion, A.; Liao, J.; Asghar, M.N.; Zhang, H.; Azhar, S.; Omary, M.B.; et al. Keratin 8 absence down-regulates colonocyte HMGCS2 and modulates colonic ketogenesis and energy metabolism. Mol. Biol. Cell 2015, 26, 2298–2310. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.M.; Sikaneta, T.; Sullivan, B.M.; Zhang, Q.; Andreucci, M.; Stehle, T.; Drummond, I.; Arnaout, M.A. Polycystin-1 interacts with intermediate filaments. J. Biol. Chem. 2001, 276, 46544–46552. [Google Scholar] [CrossRef] [PubMed]
- Basora, N.; Tetreault, M.P.; Boucher, M.P.; Herring, E.; Beaulieu, J.F. Polycystin-1 is a microtubule-driven desmosome-associated component in polarized epithelial cells. Exp. Cell Res. 2010, 316, 1454–1464. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Sun, Y.; Zhang, F.; Zhang, W.K.; Wang, D.; Wang, Y.; Cao, X.; Hu, W.; Xie, C.; Cuppoletti, J.; et al. Keratin K18 increases cystic fibrosis transmembrane conductance regulator (CFTR) surface expression by binding to its C-terminal hydrophobic patch. J. Biol. Chem. 2012, 287, 40547–40559. [Google Scholar] [CrossRef] [PubMed]
- Nataro, J.P.; Kaper, J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998, 11, 142–201. [Google Scholar] [PubMed]
- Frankel, G.; Phillips, A.D.; Rosenshine, I.; Dougan, G.; Kaper, J.B.; Knutton, S. Enteropathogenic and enterohaemorrhagic Escherichia coli: More subversive elements. Mol. Microbiol. 1998, 30, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Rothbaum, R.; McAdams, A.J.; Giannella, R.; Partin, J.C. A clinicopathologic study of enterocyte-adherent Escherichia coli: A cause of protracted diarrhea in infants. Gastroenterology 1982, 83, 441–454. [Google Scholar] [PubMed]
- Bement, W.M.; Mooseker, M.S. The cytoskeleton of the intestinal epithelium: Components, assembly, and dynamic rearrangements. In The Cytoskeleton: A Multi-Volume Treatise; John, E.H., Ian, F.P., Eds.; JAI Press: Greenwich, CT, USA, 1996; Volume 3, pp. 359–404. [Google Scholar]
- Gruenheid, S.; DeVinney, R.; Bladt, F.; Goosney, D.; Gelkop, S.; Gish, G.D.; Pawson, T.; Finlay, B.B. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 2001, 3, 856–859. [Google Scholar] [CrossRef] [PubMed]
- Rowe, B.; Ward, L.R.; Threlfall, E.J. Multidrug-resistant Salmonella typhi: A worldwide epidemic. Clin. Infect. Dis. 1997, 24 (Suppl. S1), S106–S109. [Google Scholar] [CrossRef] [PubMed]
- Ginocchio, C.; Pace, J.; Galan, J.E. Identification and molecular characterization of a Salmonella typhimurium gene involved in triggering the internalization of Salmonellae into cultured epithelial cells. Proc. Natl. Acad. Sci. USA 1992, 89, 5976–5980. [Google Scholar] [CrossRef] [PubMed]
- Francis, C.L.; Starnbach, M.N.; Falkow, S. Morphological and cytoskeletal changes in epithelial cells occur immediately upon interaction with Salmonella typhimurium grown under low-oxygen conditions. Mol. Microbiol. 1992, 6, 3077–3087. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.D.; Paterson, H.F.; Hall, A.; Falkow, S. Salmonella typhimurium induces membrane ruffling by a growth factor-receptor-independent mechanism. Proc. Natl. Acad. Sci. USA 1993, 90, 10390–10394. [Google Scholar] [CrossRef] [PubMed]
- Finlay, B.B.; Ruschkowski, S.; Dedhar, S. Cytoskeletal rearrangements accompanying Salmonella entry into epithelial cells. J. Cell Sci. 1991, 99, 283–296. [Google Scholar] [PubMed]
- Garcia-del Portillo, F.; Pucciarelli, M.G.; Jefferies, W.A.; Finlay, B.B. Salmonella typhimurium induces selective aggregation and internalization of host cell surface proteins during invasion of epithelial cells. J. Cell Sci. 1994, 107, 2005–2020. [Google Scholar] [PubMed]
- Kaniga, K.; Tucker, S.; Trollinger, D.; Galan, J.E. Homologs of the shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 1995, 177, 3965–3971. [Google Scholar] [PubMed]
- Gebert, A.; Rothkotter, H.J.; Pabst, R. Cytokeratin 18 is an M-cell marker in porcine Peyer’s patches. Cell Tissue Res. 1994, 276, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.D.; Ghori, N.; Falkow, S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J. Exp. Med. 1994, 180, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Galan, J.E.; Lara-Tejero, M.; Marlovits, T.C.; Wagner, S. Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 2014, 68, 415–438. [Google Scholar] [CrossRef] [PubMed]
- Bennish, M.L. Potentially lethal complications of shigellosis. Rev. Infect. Dis. 1991, 13 (Suppl. S4), S319–S324. [Google Scholar] [CrossRef] [PubMed]
- Jennison, A.V.; Verma, N.K. Shigella flexneri infection: Pathogenesis and vaccine development. FEMS Microbiol. Rev. 2004, 28, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Henderson, I.R.; Navarro-Garcia, F.; Desvaux, M.; Fernandez, R.C.; Ala’Aldeen, D. Type V protein secretion pathway: The autotransporter story. Microbiol. Mol. Biol. Rev. 2004, 68, 692–744. [Google Scholar] [CrossRef] [PubMed]
- Pallen, M.J.; Chaudhuri, R.R.; Henderson, I.R. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 2003, 6, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Sun, S.; Liu, Y.; Li, J.; Zhang, T.; Wu, H.; Chen, X.; Chen, D.; Zhou, Y. Keratin 18 phosphorylation as a progression marker of chronic hepatitis B. Virol. J. 2010, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Toivola, D.M.; Ku, N.O.; Resurreccion, E.Z.; Nelson, D.R.; Wright, T.L.; Omary, M.B. Keratin 8 and 18 hyperphosphorylation is a marker of progression of human liver disease. Hepatology 2004, 40, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Toivola, D.; Ostrowski, S.; Baribault, H.; Magin, T.; Ramsingh, A.; Omary, M. Keratins provide virus-dependent protection or predisposition to injury in coxsackievirus-induced pancreatitis. Cell Health Cytoskelet. 2009, 1, 51–65. [Google Scholar]
- Kumar, Y.; Valdivia, R.H. Actin and intermediate filaments stabilize the chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe 2008, 4, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Valdivia, R.H. Reorganization of the host cytoskeleton by the intracellular pathogen chlamydia trachomatis. Commun. Integr. Biol. 2008, 1, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Mattos, E.C.; Tonelli, R.R.; Colli, W.; Alves, M.J. The gp85 surface glycoproteins from Trypanosoma cruzi. Subcell Biochem. 2014, 74, 151–180. [Google Scholar] [PubMed]
- Markl, J.; Franke, W.W. Localization of cytokeratins in tissues of the rainbow trout: Fundamental differences in expression pattern between fish and higher vertebrates. Differentiation 1988, 39, 97–122. [Google Scholar] [CrossRef] [PubMed]
- Maurizii, M.G.; Alibardi, L.; Taddei, C. Organization and characterization of the keratin cytoskeleton in the previtellogenic ovarian follicle of the lizard Podarcis sicula raf. Mol. Reprod. Dev. 2000, 57, 159–166. [Google Scholar] [CrossRef]
- Bossinger, O.; Fukushige, T.; Claeys, M.; Borgonie, G.; McGhee, J.D. The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by let-413. Dev. Biol. 2004, 268, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Carberry, K.; Wiesenfahrt, T.; Windoffer, R.; Bossinger, O.; Leube, R.E. Intermediate filaments in caenorhabditis elegans. Cell Motil. Cytoskelet. 2009, 66, 852–864. [Google Scholar] [CrossRef] [PubMed]
- Jahnel, O.; Hoffmann, B.; Merkel, R.; Bossinger, O.; Leube, R.E. Mechanical probing of the intermediate filament-rich Caenorhabditis elegans intestine. Methods Enzymol. 2016, 568, 681–706. [Google Scholar] [PubMed]
- Hüsken, K.; Wiesenfahrt, T.; Abraham, C.; Windoffer, R.; Bossinger, O.; Leube, R.E. Maintenance of the intestinal tube in Caenorhabditis elegans: The role of the intermediate filament protein IFC-2. Differentiation 2008, 76, 881–896. [Google Scholar] [CrossRef] [PubMed]
- Carberry, K.; Wiesenfahrt, T.; Geisler, F.; Stocker, S.; Gerhardus, H.; Uberbach, D.; Davis, W.; Jorgensen, E.; Leube, R.E.; Bossinger, O. The novel intestinal filament organizer IFO-1 contributes to epithelial integrity in concert with ERM-1 and DLG-1. Development 2012, 139, 1851–1862. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.Y.; Los, F.C.; Huffman, D.L.; Wachi, S.; Kloft, N.; Husmann, M.; Karabrahimi, V.; Schwartz, J.L.; Bellier, A.; Ha, C.; et al. Global functional analyses of cellular responses to pore-forming toxins. PLoS Pathog. 2011, 7, e1001314. [Google Scholar] [CrossRef] [PubMed]
- Estes, K.A.; Szumowski, S.C.; Troemel, E.R. Non-lytic, actin-based exit of intracellular parasites from C. elegans intestinal cells. PLoS Pathog 2011, 7, e1002227. [Google Scholar] [CrossRef] [PubMed]
- Stutz, K.; Kaech, A.; Aebi, M.; Kunzler, M.; Hengartner, M.O. Disruption of the C. elegans intestinal brush border by the fungal lectin CCL2 phenocopies dietary lectin toxicity in mammals. PLoS ONE 2015, 10, e0129381. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, B.; Gonzalez-Rodriguez, I.; Arboleya, S.; Lopez, P.; Suarez, A.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M. The effects of bifidobacterium breve on immune mediators and proteome of HT29 cells monolayers. BioMed Res. Int. 2015, 2015, 479140. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, R.P.; Batazzi, A.S.; Han, M.C.; Coulombe, P.A. Loss of keratin 17 induces tissue-specific cytokine polarization and cellular differentiation in HPV16-driven cervical tumorigenesis in vivo. Oncogene 2016. [Google Scholar] [CrossRef] [PubMed]


| Pathogen | Mechanism | Effect | Cell Type | Reference | |
|---|---|---|---|---|---|
| Staphylococcus aureus | Staphylococcal surface protein clumping factor B (ClfB)-dependent adherence to K10 | Epithelial colonization | Squamous nasal epithelial cells | [52] | |
| Streptococcus agalactiae | Streptococcal surface-localized serine-rich repeat protein Srr-1 binding to K4 | Epithelial colonization | Saliva extracts | [53] | |
| Human papilloma virus type 16 | Association of HPV type 16 E1^E4 protein with K18 followed by K18-S33 and K18-S52 phosphorylation and ubiquitinylation | Keratin network disruption | SiHa and HaCaT cells | [54] | |
| Herpes simplex virus type 2 | Association of US2 with K18 | Keratin network disruption | Vero and A431 cells | [55] | |
| Herpes simplex virus type 2 | Association of US3 with K17 followed by keratin phosphorylation and ubiquitinylation | Keratin network disruption | Hep2 cells | [56] | |
| Porphyromonas gingivalis | Cleavage of K6 at K357-Y358 and K378-Q379 by lysine-specific gingipain | Induction of inflammation | Gingival epithelial cells | [57] | |
| Pseudomonas aeruginosa | Release of K6-derived antibacterial peptides | Bacteriotoxicity | hTCEpi cells | [58] | |
| Enteropathogenic Escherichia coli | K18-dependent actin filament reorganization | Pathogen docking | HeLa cells | [59] | |
| Salmonella enterica serovar typhimurium | Interaction of secreted Salmonella invasion protein SipC with K18 | Pathogen docking | HEp-2 cells | [60] | |
| Salmonella enterica serovar typhimurium | Interaction of Salmonella type III secretion translocon protein SspC with K8 | Pathogen docking | HeLa cells | [61] | |
| Shigella flexneri | Binding of Shigella translocon pore protein IpaC to K18 | Pathogen docking | [62] | ||
| Enterobacteriaceae | Binding of the serine protease autotransporter of Enterobacteriaceae Pet to K8 | Induction of cytotoxicity | HT-29 and HEp-2 cells | [63] | |
| Rotavirus | Phosphorylation of K8 | Keratin network disruption | HT29 cells | [64] | |
| Adenovirus | Cleavage of aminoterminal K18 head domain at position 73 | Keratin network disruption | HeLa and 293 cells | [65,66] | |
| Rhinovirus | Cleavage of aminoterminal K8 head domain at position 14 by 2A proteinase | Keratin network disruption | HeLa cell extracts | [67] | |
| Chlamydia trachomatis | Cleavage of K8 by chlamydial protease-like activity factor CPAF | Keratin network disruption | HeLa cells | [68] | |
| Chlamydia pneumoniae | Cleavage of K8 and K18 by chlamydial protease-like activity factor CPAF | Keratin network disruption | HL cells | [69] | |
| Trypanosoma cruzi | Binding of peptide TS9 of glycoprotein gp85 to K8/K18 (K14, K19, K20) | Cytoplasmic proliferation | LLC-MK2 cell extract | [70] | |
| Spraguea lophii | Phosphorylation of K4 and K13 in the outer spore envelope | Polar tube release | [71] |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geisler, F.; Leube, R.E. Epithelial Intermediate Filaments: Guardians against Microbial Infection? Cells 2016, 5, 29. https://doi.org/10.3390/cells5030029
Geisler F, Leube RE. Epithelial Intermediate Filaments: Guardians against Microbial Infection? Cells. 2016; 5(3):29. https://doi.org/10.3390/cells5030029
Chicago/Turabian StyleGeisler, Florian, and Rudolf E. Leube. 2016. "Epithelial Intermediate Filaments: Guardians against Microbial Infection?" Cells 5, no. 3: 29. https://doi.org/10.3390/cells5030029
APA StyleGeisler, F., & Leube, R. E. (2016). Epithelial Intermediate Filaments: Guardians against Microbial Infection? Cells, 5(3), 29. https://doi.org/10.3390/cells5030029